The approach described in previous sections relies on ANOVA to detect differentially methylated CpG sites and takes individual sites as a starting point for interpretation. Since ANOVA compares M values at each site independently, this strategy is robust to type I/type II probe bias.
An alternative could be to first summarize all the probes belonging to a CpG island region (i.e. island, N-shore, N-shelf, S-shore, S-shelf) and then use ANOVA to compare regions across the groups. Since the summarization will include both type I and type II probes, you may want to split the analysis in two branches and analyze type I and type II probes independently. To do this, we need to annotate each probe as type I or type II.
- Select the mvalue spreadsheet
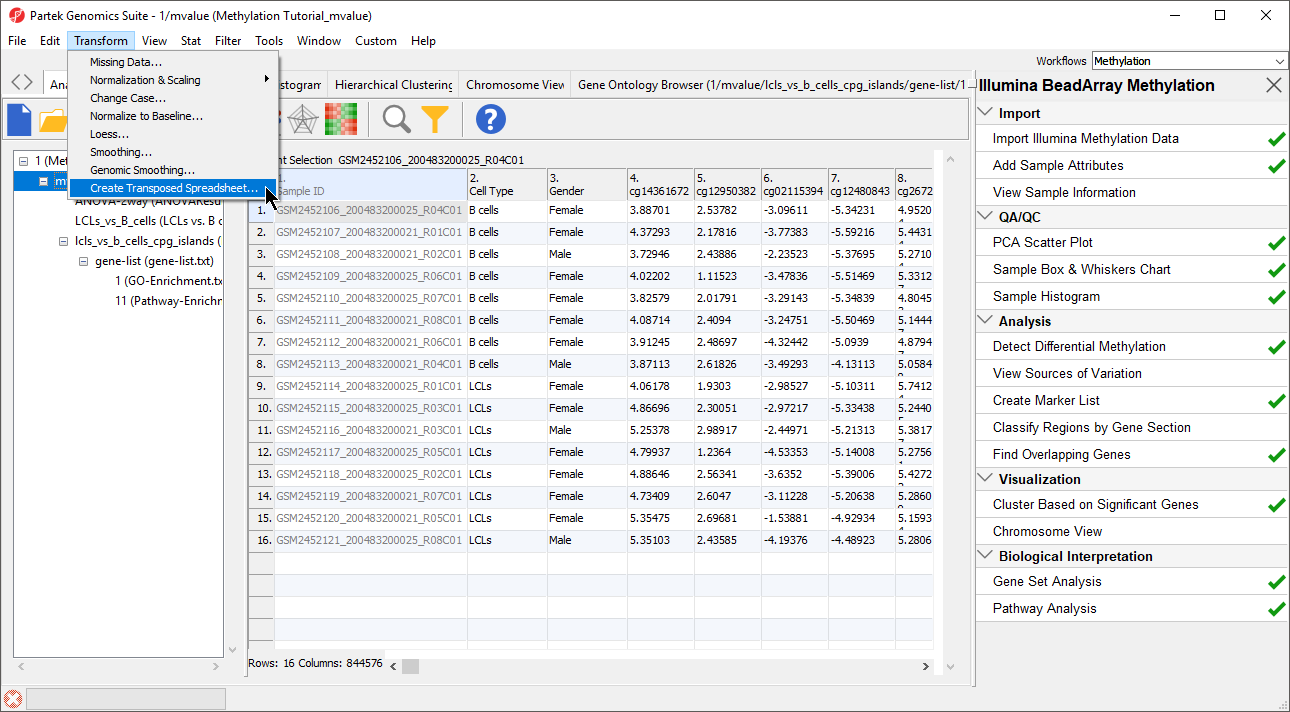
- Select Transform from the main toolbar
- Select Create Transposed Spreadsheet... from the Transform drop-down menu (Figure 1)
- Select Sample ID for Column: and numeric for Data Type:
- Select OK
A new temporary spreadsheet will be created with a row for each probe and columns for each sample.
- Right-click on column 1. ID to bring up the pop-up menu
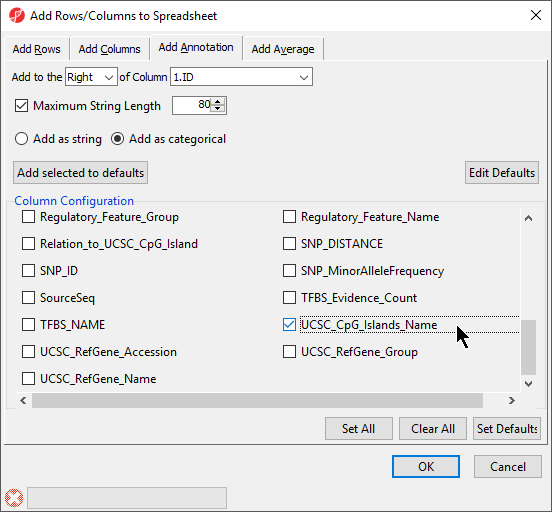
- Select Insert Annotation
- Select Add as categorical
- Select Infinium_Design_Type and UCSC_CpG_Islands_Name from the Column Configuration options (Figure 2)
- Select OK to add the Inifinium design type and UCSC CpG island name as categorical columns on the spreadsheet
Now, we can use the interactive filter to create separate spreadsheets for type I and type II probes.
- Select () to launch the interactive filter
- Select 2. Infinium_Design_Type from the drop-down menu if not selected by default
- Left-click the type I column to exclude it
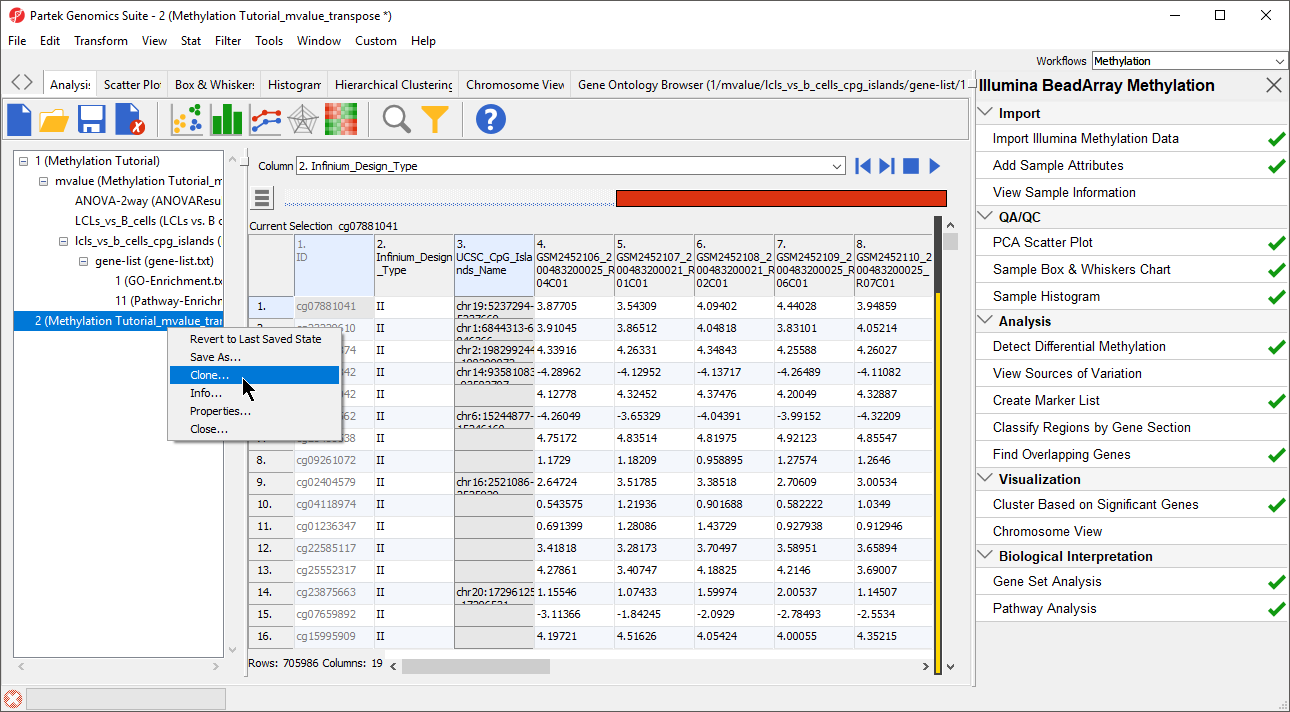
- Right-click the temporary spreadsheet in the spreadsheet tree to bring up the pop-up dialog
- Select Clone... (Figure 3)
- Name the new spreadsheet female_only_typeII_probes
- Select OK
- Save the created spreadsheet, we chose the file name female_only_typeII_probes
- Repeat process to create a spreadsheet for type I probes
The temporary spreadsheet is no longer needed so we can close it.
- Close the temporary spreadsheet by selecting it in the file tree and selecting ()
We can use these spreadsheets to generate lists of M values at CpG island regions
- Select spreadsheet female_only_typeII_probes
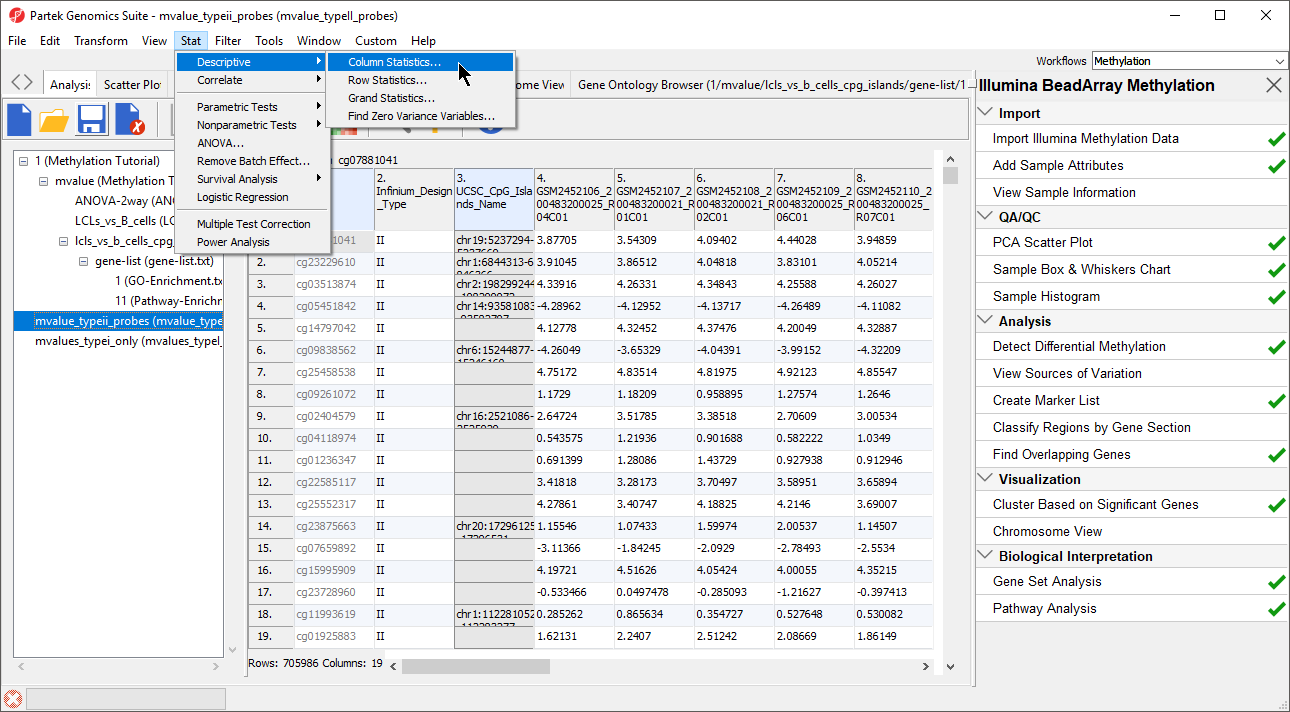
- Select Stat from the main toolbar
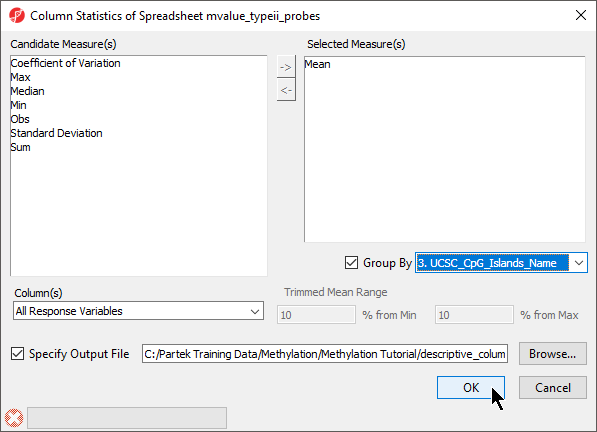
- Select Column Statistics... under Descriptive (Figure 4)
- Add Mean to the Selected Measure(s) panel
- Select Group By and set it to 3. UCSC_CpG_Islands_Name (Figure 5)
- Select OK
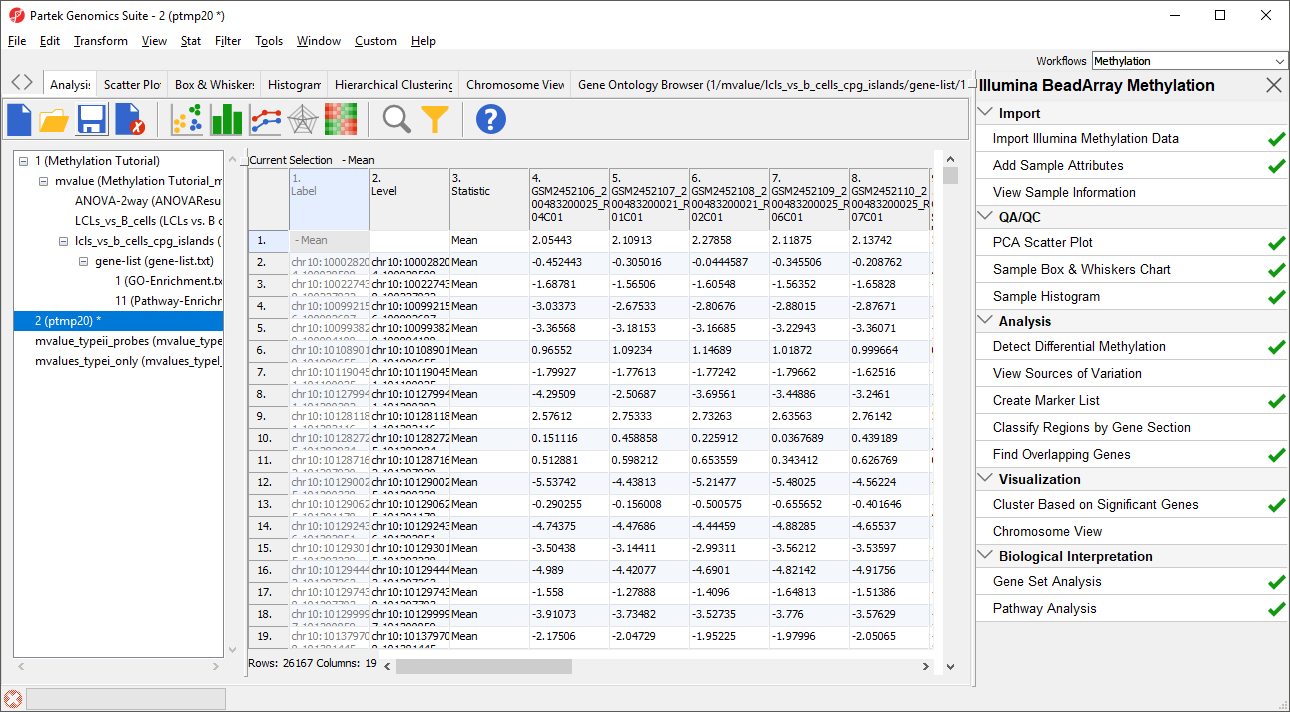
The new temporary spreadsheet has one CpG island region per row (Figure 6), samples on columns, and the values in the cells represent the mean of M values of all the CpG probes in the region.
Note the first row, with label “– Mean”. It corresponds to all the probes that map outside of UCSC CpG islands. As it is not needed for the downstream analysis, we will remove it.
- Right-click on the row header for Mean
- Select Delete to remove the row
The final step is to transpose the data back to its original orientation.
- Select Transform from the main toolbar
- Select Create Transposed Spreadsheet... from the Transform drop-down menu
- Select 2. Level for Column: and numeric for Data Type:
- Select OK
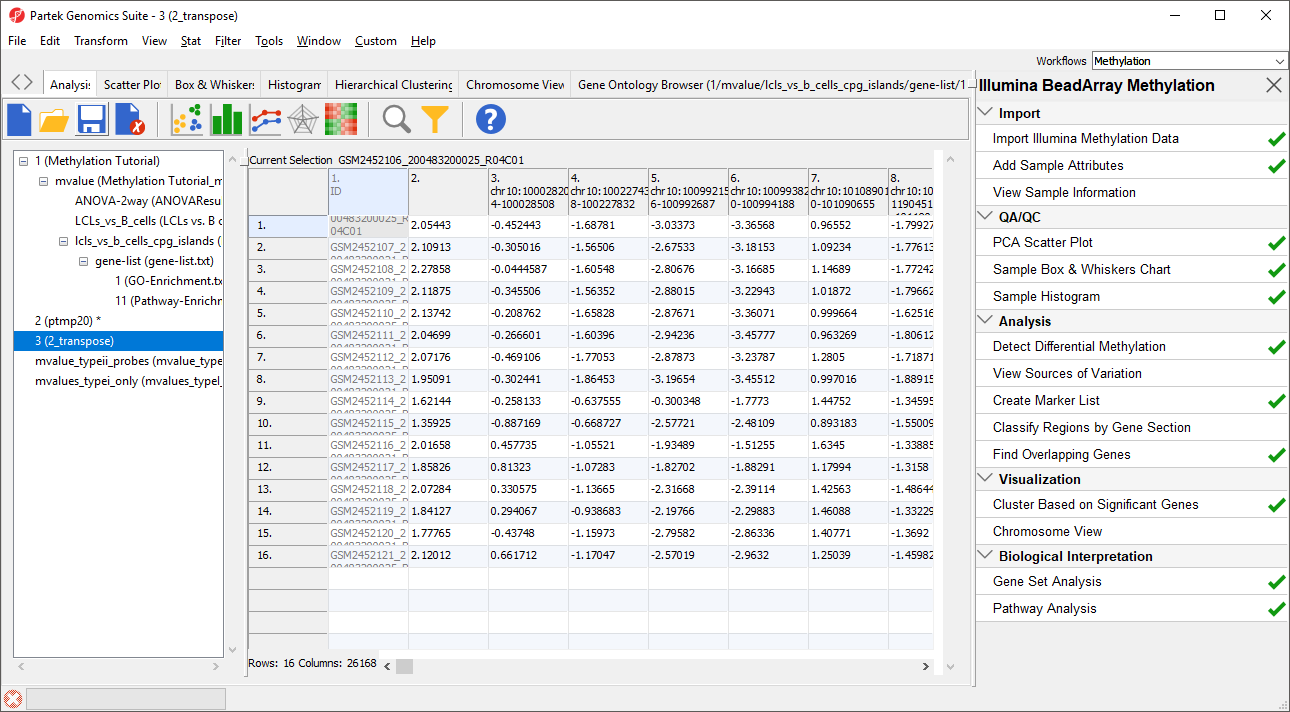
The layout of the new transposed spreadsheet is as follows: one sample per row with CpG island regions on columns; cell entries correspond to mean methylation status of the region (Figure 7). The column with a blank value for the column header is the average of all probes not associated with CpG island regions. You can delete this column if you like.
- Right-click the transposed spreadsheet, 2_transpose
- Select Save as... from the pop-up menu
- Name it mvalues_typeII_probes_CpG_islands
- Close the source temporary spreadsheet by selecting it in the spreadsheet tree and selecting ()
The mvalues_typeII_probes_CpG_islands spreadsheet can be used as a starting point for ANOVA and other analyses. You can also repeat the steps above to create an equivalent spreadsheet for type I probes.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |