The basic method of creating a gene list from ANOVA results based on fold-change and p-value cut-offs is detailed in Creating gene lists from ANOVA results. Advanced options enable the creation of lists based on more complex criteria. For example, we can use the Create Gene List function to identify transcripts that are both significantly differentially expressed AND alternatively-spliced among the four tissue samples.
- Select Create Gene List from the Analyze Known Genes panel of the RNA-Seq workflow to invoke the List Manager dialog
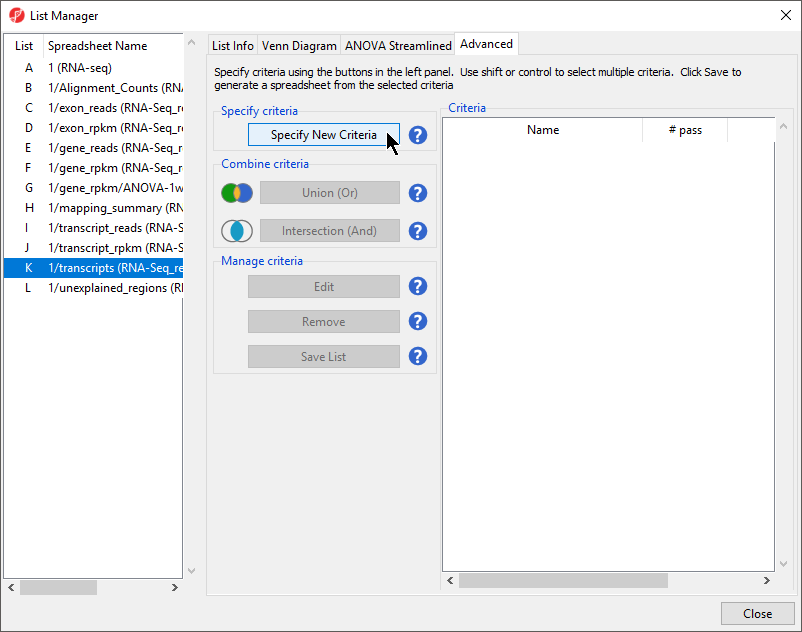
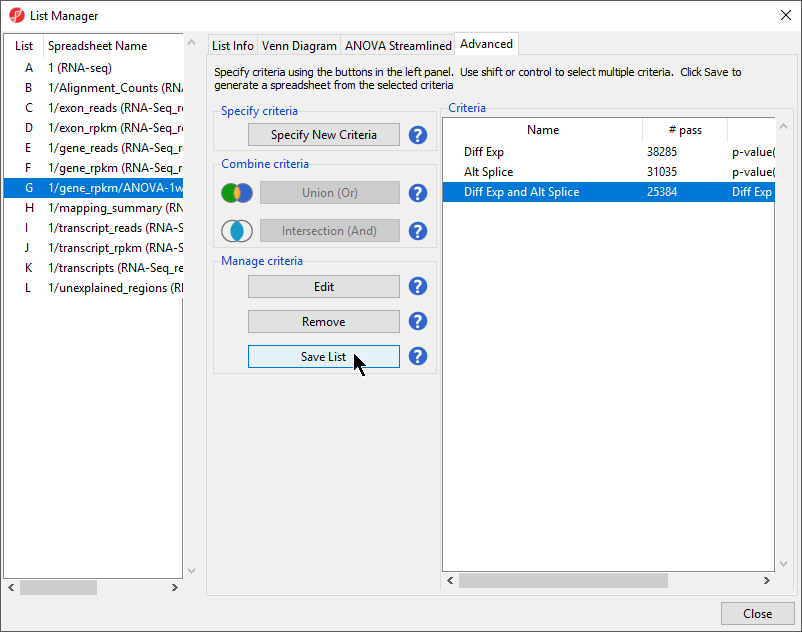
- Select the Advanced tab (Figure 1)
- Select Specify New Criteria to invoke the Configure Criteria dialog (Figure 2)
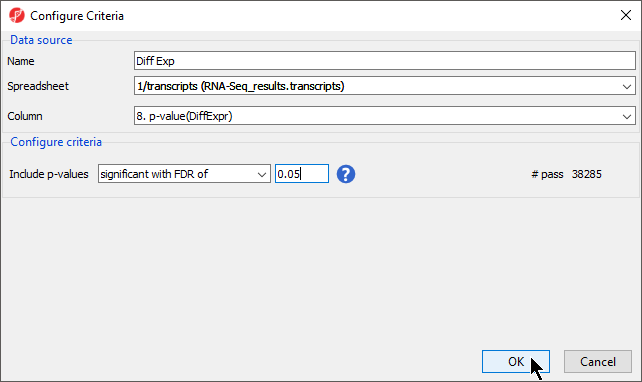
- In the Configure Criteria dialog box (Figure 2), provide a name for the list (Diff Exp)
- Select 1/transcripts (RNA-Seq_results.transcripts) from theSpreadsheet drop-down menu
- Select 8. p-value(DiffExp) from the Column drop-down menu
- Set Include p-values to significant with FDR with a value of 0.05
A list of 38,285 transcripts that pass this criteria will be generated according to the # pass score on the right-hand side of the dialog. If the settings are changed, this number will automatically update.
- Select OK
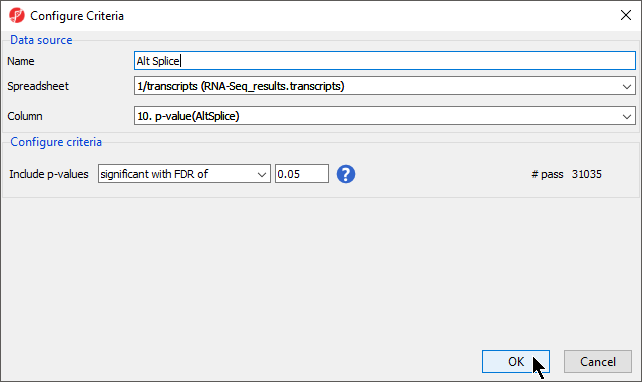
- Repeat the same steps to create a list of transcripts that are likely alternatively spliced, named Alt Splice, using the same p-value cutoff and Column set to 10. p-value (AltSplice) (Figure 3)
- Select OK to generate Alt Splice
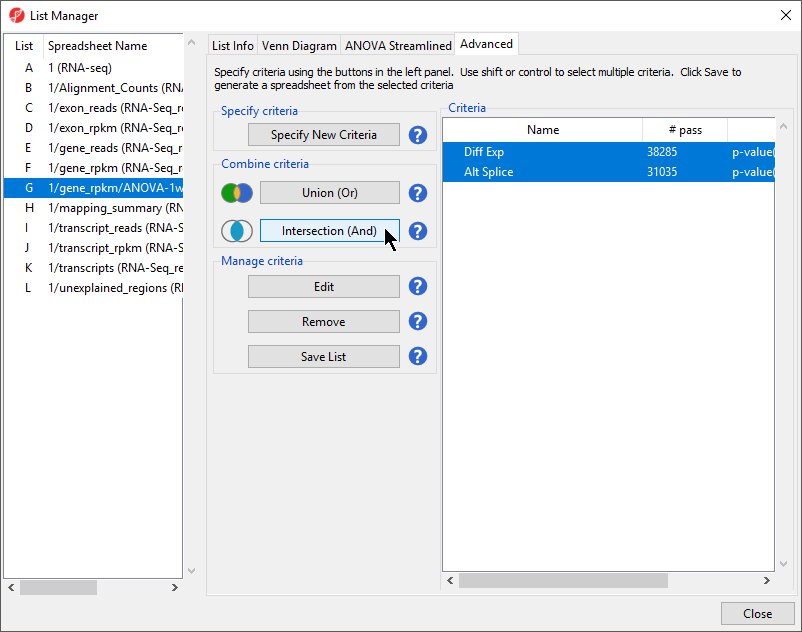
- Select both lists in the right-hand panel under the Criteria panel while holding the Ctrl key on your keyboard
- Select Intersection from the left-hand panel of the List Manager dialog (Figure 4)
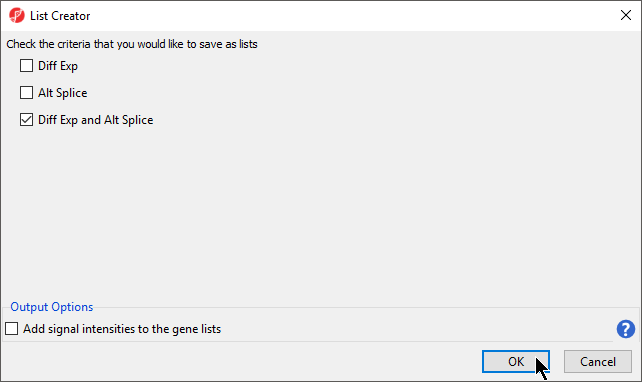
- Enter a name for the criteria (Diff Exp and Alt Splice)
- Select OK to close the naming dialog and OK again to close the list creation hint dialog
- Select Save List from the Manage criteria section of the List Manager dialog (Figure 5)
- Select Diff Exp and Alt Splice in the List Creator dialog (Figure 6)
- Select OK to save the list
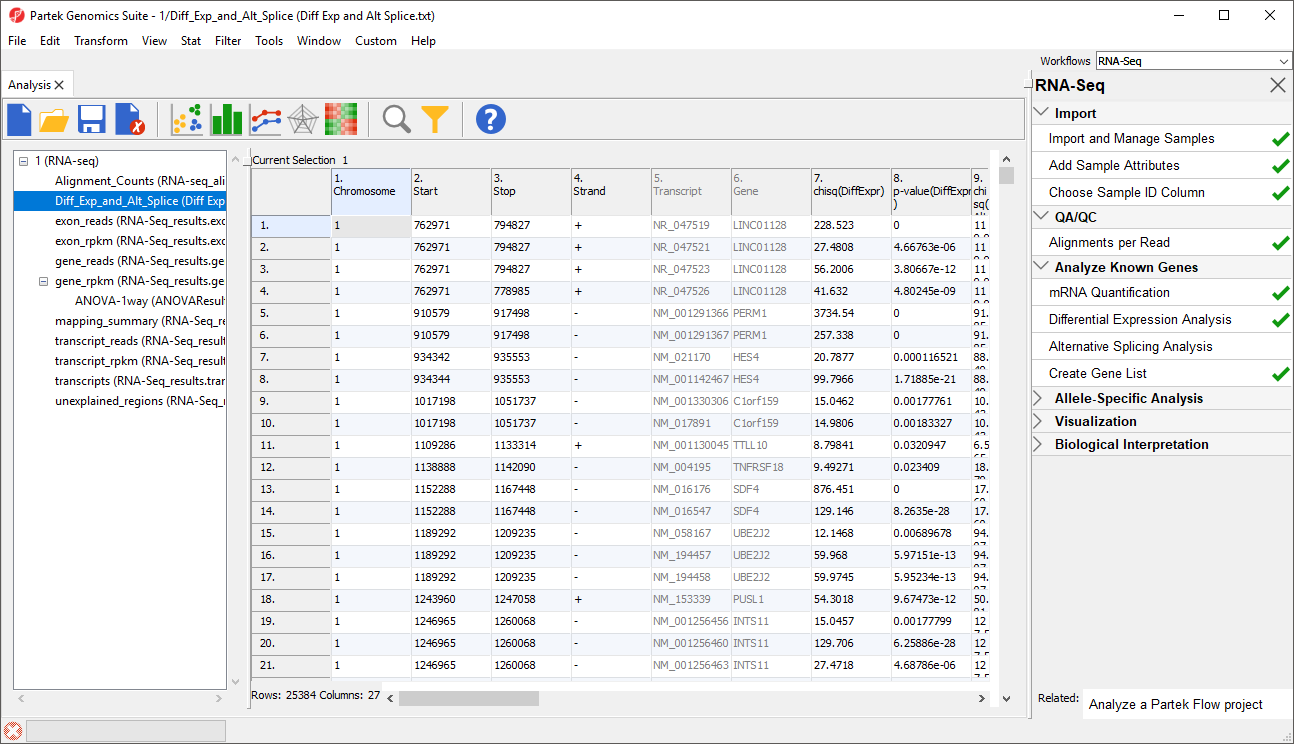
- Select Close to exit the List Manager dialog and view the Diff_Exp_and_Alt_Splice spreadsheet (Figure 7)
This list of differentially expressed and alternatively spliced transcripts will be used later in the tutorial.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |