The first step in analyzing Affymetrix intensity data is to estimate the number of copies of each marker (allele).
- Select Create Copy Number (from Allele Intensities Only)
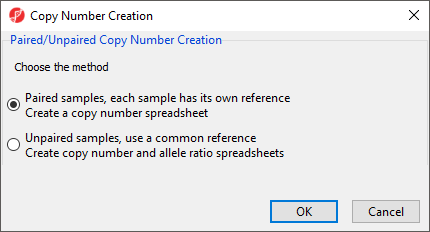
This launches the Copy Number Creation dialog (Figure 1).
Choosing Paired samples assumes that each sample has its own reference sample with a common sample ID and generates a copy number spreadsheet. Choosing Unpaired samples uses a common reference, either a single sample or a group of samples, to create both a copy number spreadsheet and an allele ratio spreadsheet.
Create Copy Number from Pairs
In this tutorial, we have paired tumor-normal samples and thus can use the Paired samples option.
- Select Paired samples
- Select OK
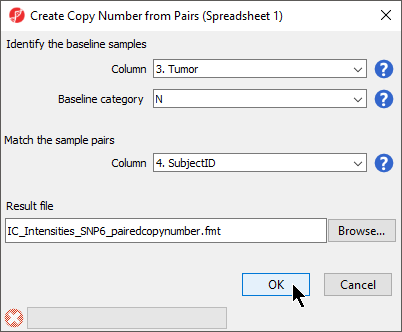
The next dialog, Create Copy Number from Pairs, asks you to choose the column shared by each pair and the column that identifies the baseline category (Figure 2).
- Select 3. Tumor for Column
- Select N for Baseline category
- Select 4. SubjectID for the Column to match sample pairs
- Select OK
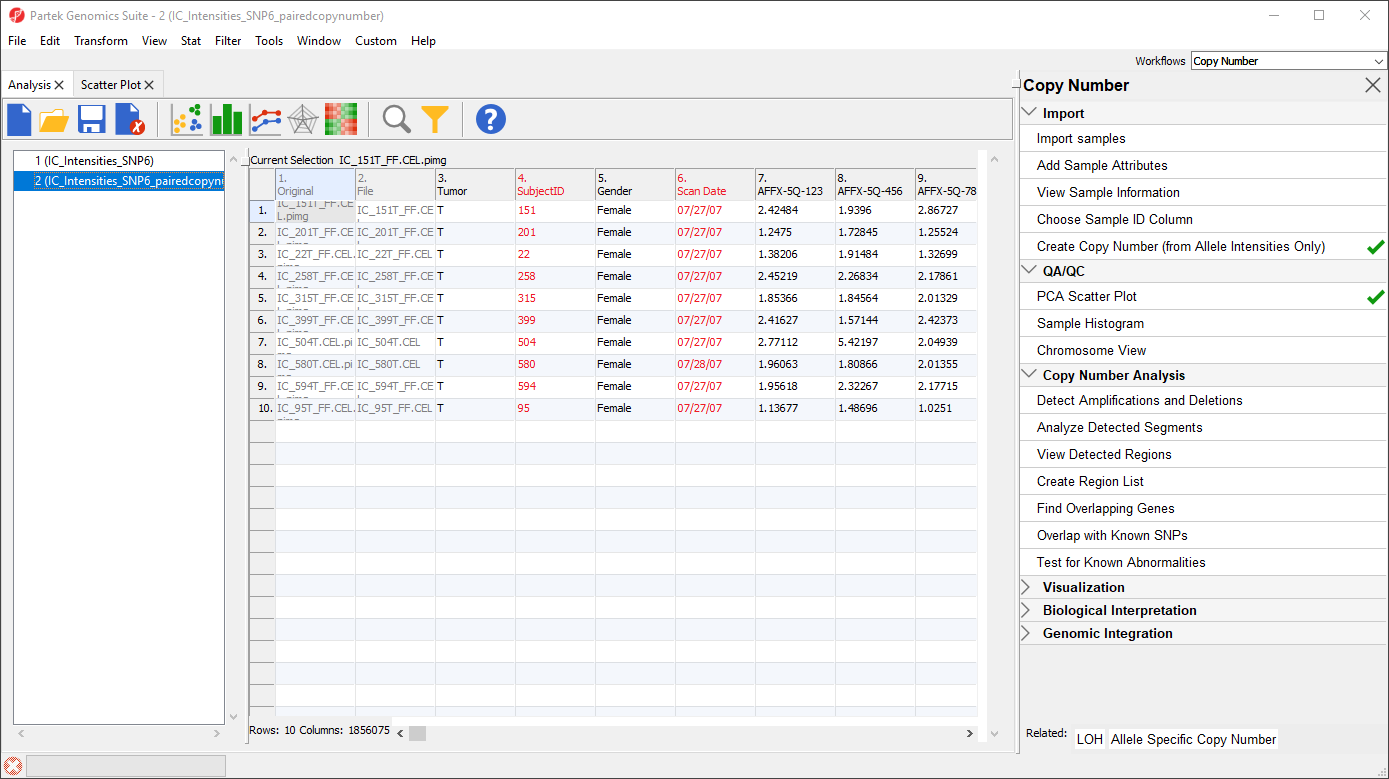
This will pair samples based on 4. SubjectID, and set the baseline sample as the sample in the pair with a value of N in the 3. Tumor column. The spreadsheet produced (Figure 3) has a row for each tumor sample. In this tutorial, columns 7+ include copy number estimates for each marker. Column 1-6 are identical to the source spreadsheet.
Create Copy Number from Reference Baseline
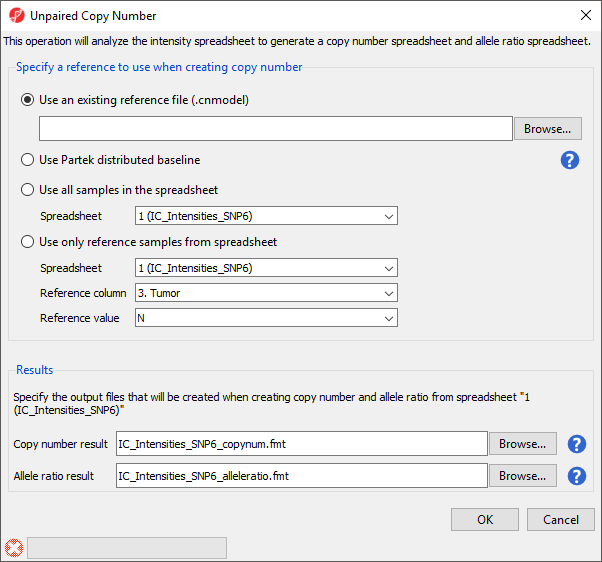
Alternatively, if paired samples are not available or appropriate, the Unpaired samples option can be selected in the Copy Number Creation dialog (Figure 1). Selecting this option opens the Unpaired Copy Number dialog (Figure 4).
There are several options for creating a reference baseline. First, you can use an existing reference file. These may be distributed by the manufacturer of your array, such as Affymetrix or Illumina, or previously created using Partek Genomics Suite from a set of normal samples. Second, you can use the reference file distributed by Partek. Third, you can choose all the samples from a separately imported spreadsheet. Fourth, you can choose a subset of the samples within the current spreadsheet to pool to create a reference.
In each case, every sample in your spreadsheet will be compared to the referece and two spreadsheets will be generated, a copy number spreadsheet and an allele ratio spreadsheet.
For more information about using unpaired samples in copy number calculations, please consult our Unpaired Copy Number Estimation white paper.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |