Because different samples have different total numbers of reads, it would be misleading to calculate differential expression by comparing read count numbers for genes across samples without normalizing for the total number of reads.
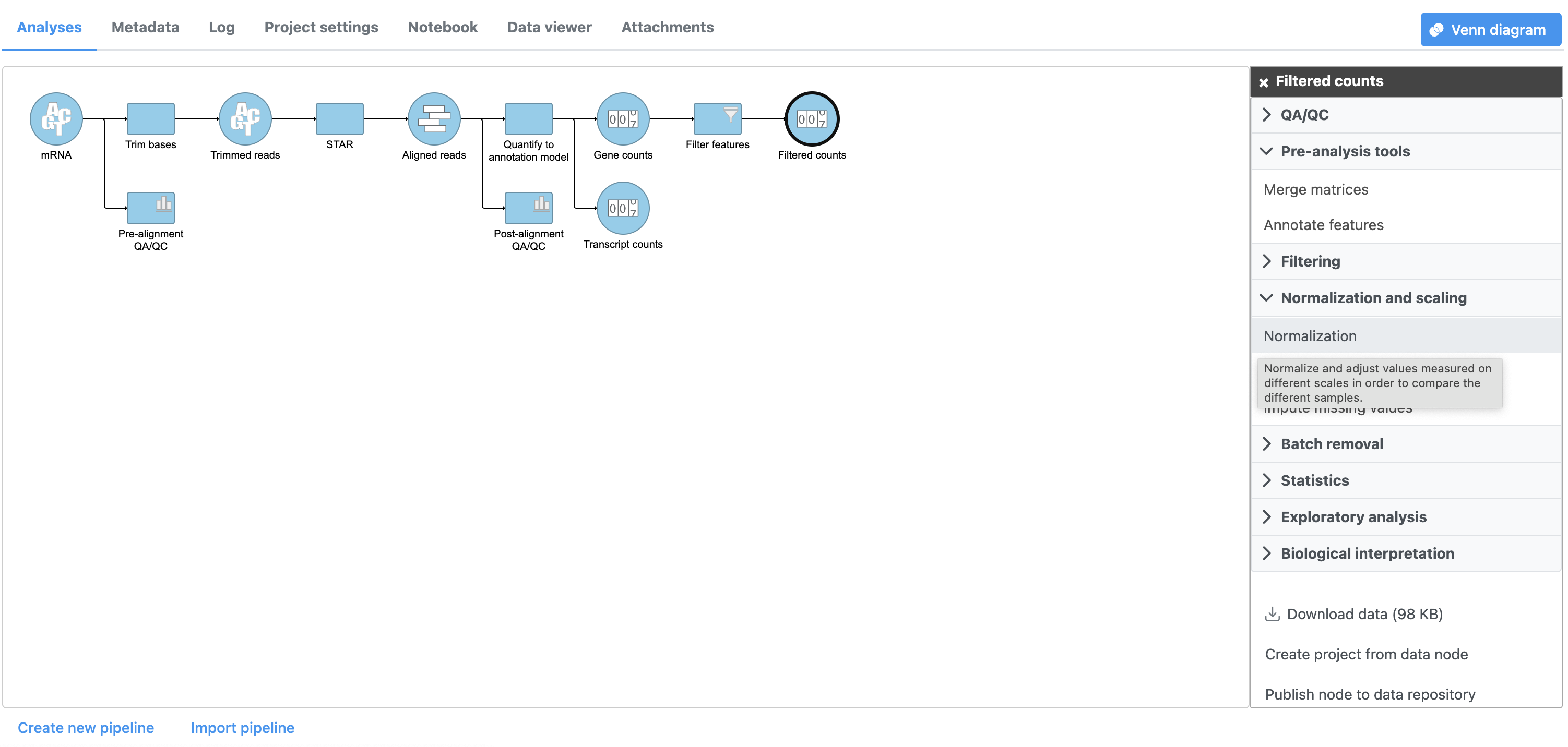
- Click the Filtered counts data node
- Click Normalization and scaling in the task menu
- Click Normalization (Figure 1)
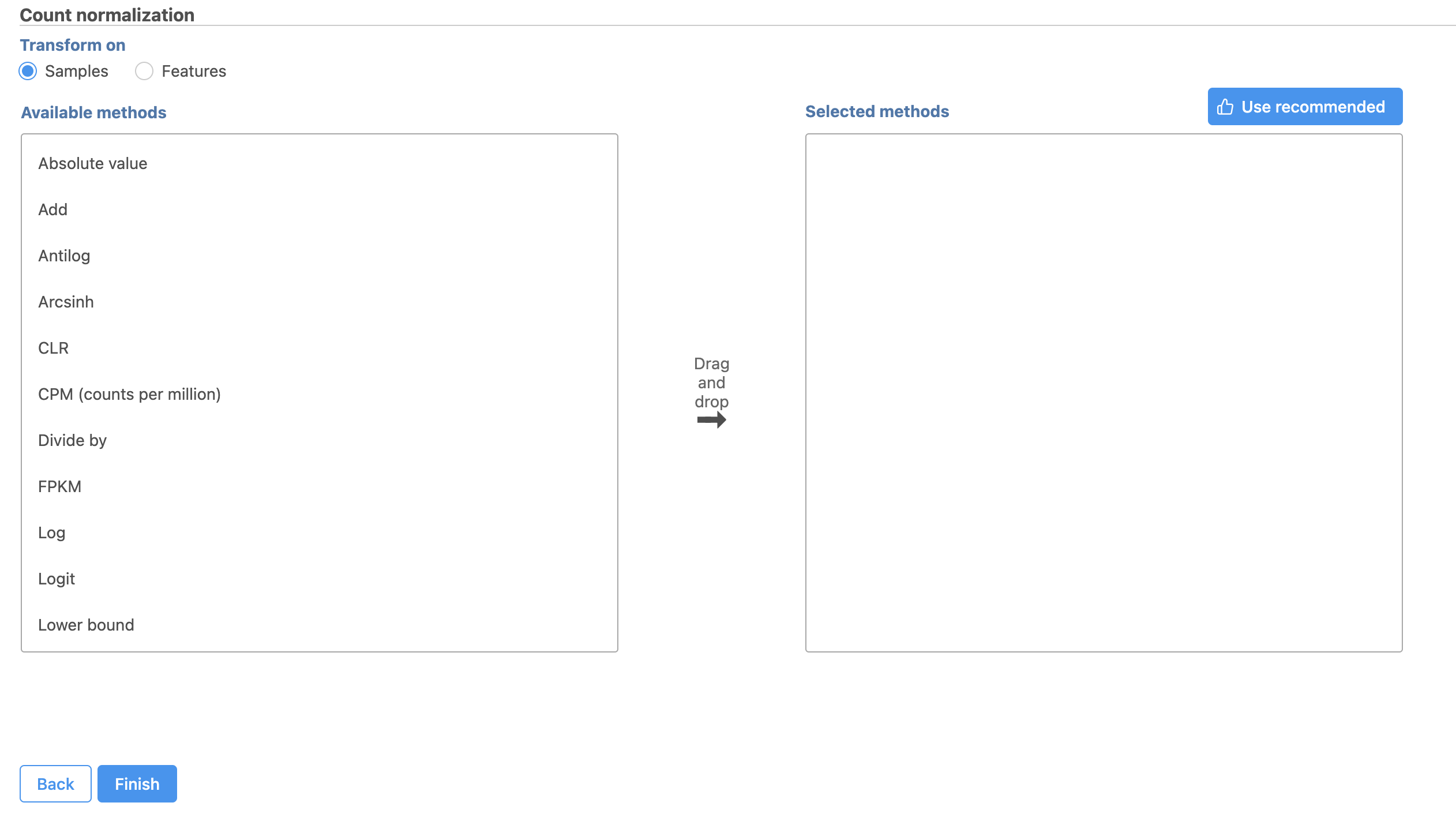
The Count normalization menu will open (Figure 2).
Normalization can be performed by sample or by feature. By sample is selected by default; this is appropriate for the tutorial data set.
Available normalization methods are listed in the left-hand panel. For more information about these options, please see the Normalize counts user guide.
For this tutorial, we will use the recommended default normalization settings.
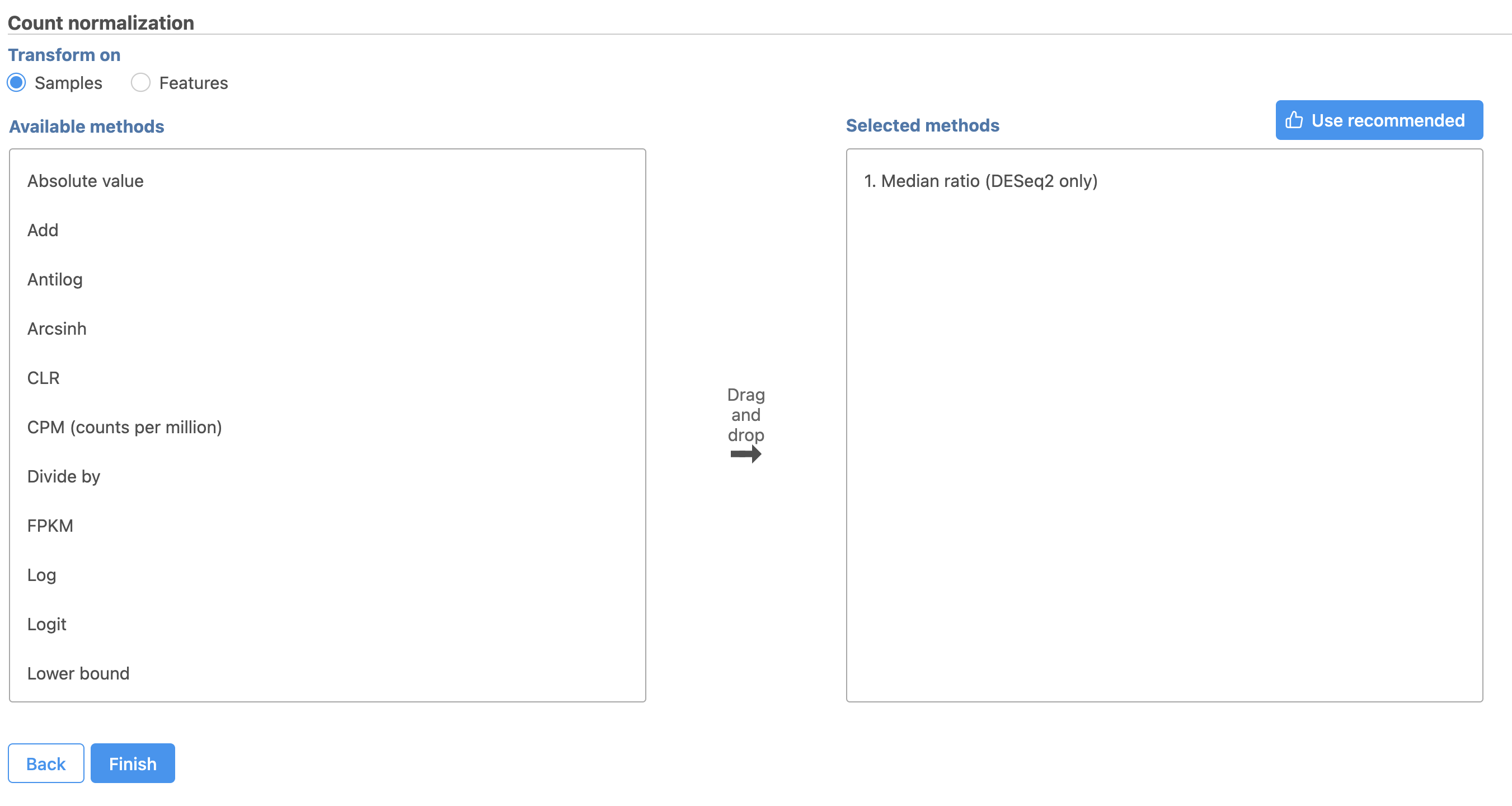
- Select
This adds the Median ratio normalization method, which is suitable for performing differential expression analysis using DESeq2 (Figure 3).
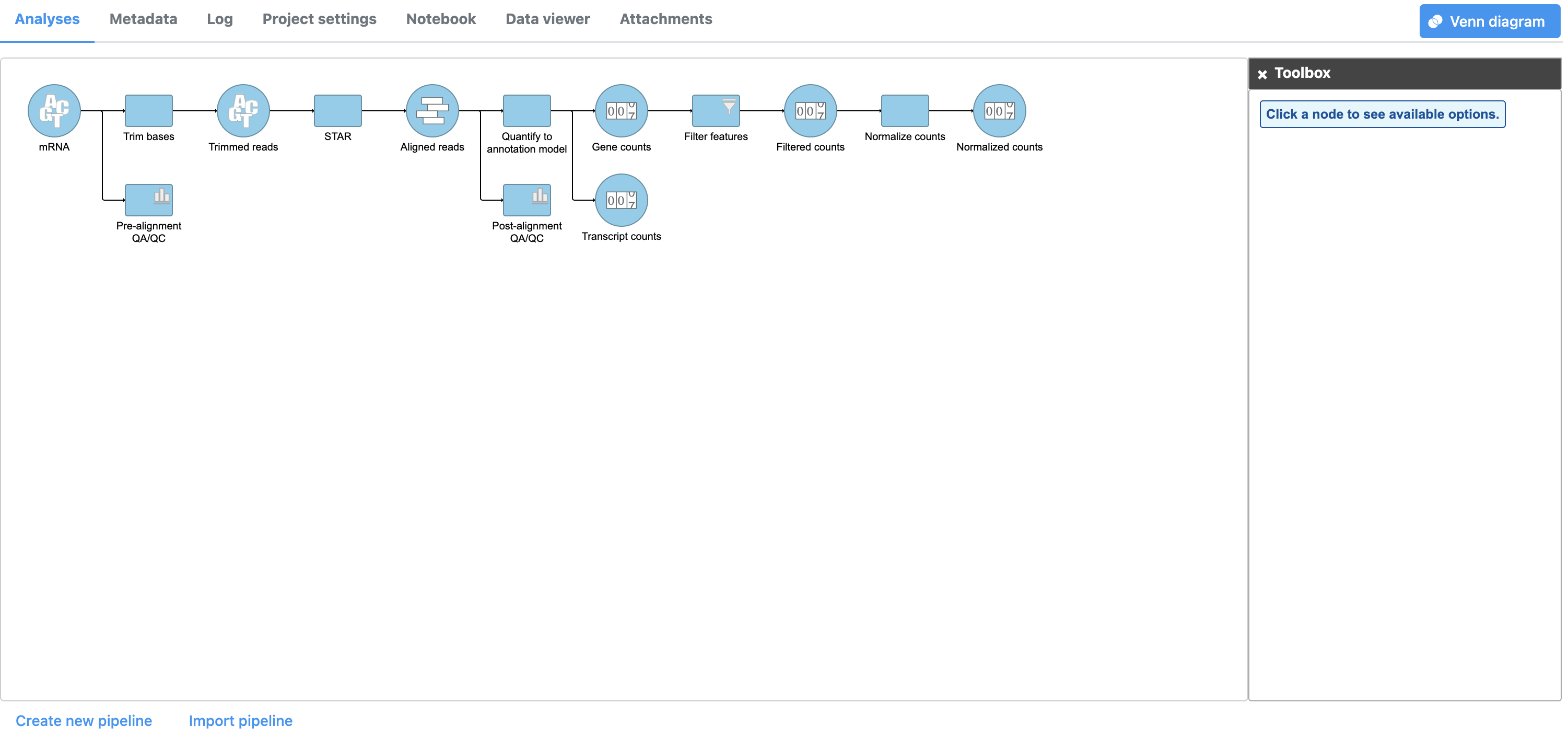
- Click Finish to perform normalization
A Normalize counts task node and a Normalized counts data node are added to the pipeline (Figure 4)
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
30 | rates |