Running a Chromosome View Task from a Data Node
On the Analysis tab, selecting a data node containing aligned reads, variants, or feature lists, shows Chromosome view in the Exploratory analysis section of the context-sensitive menu (Figure 1).
If Partek Flow has no information on the genome build, you will need to provide the species and genome build in a subsequent dialog (not shown). Otherwise, chromosome view will come up directly.
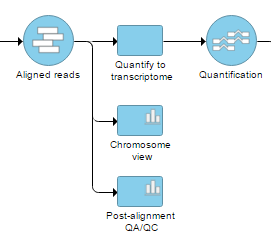
A new Chromosome view task node will be added to the canvas (Figure 2) and in order to invoke the viewer <double-click> on the node (you can also select it and then go to Task report in the menu). When invoked in this way, the default visualization in the Chromosome view is the first 100,000 bases of the first chromosome.
Browsing directly to a location directly from a Task Report
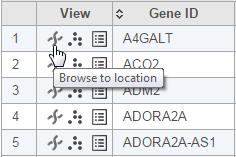
Another way to get the Chromosome view is through a Task report. You can launch the viewer by selecting the chromosome icon in the View column (Figure 3) of GSA or View Variants reports. In that case, the Chromosome view will browse directly to the selected genomic location (i.e. a transcript or a variant, depending on the pipeline).
Depending on the task report used to invoke the Chromosome view was invoked, some tracks may be pre-selected and customized. For example, when invoked from a variant table, the reads histogram track will be colored by bases (rather than the default of coloring by sample).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
35 | rates |

1 Comment
Melissa del Rosario
author: ilukic