Controls
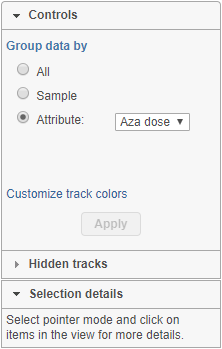
Chromosome view can be customized by using the control panel on the left (Figure 1). The Attribute and Order By controls show options depending on the current project, while the content of the Annotate amino acids control depends on the annotation files associated with the current genome build in the Library File Management. In order for any change to take place, push the Apply button.
Group data by
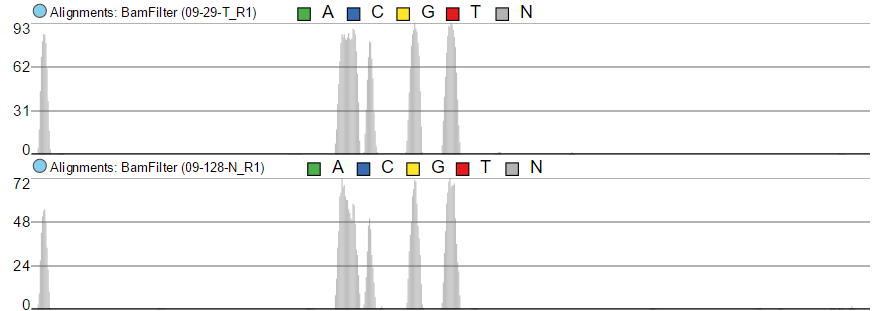
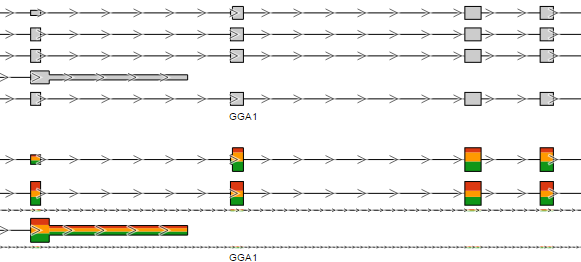
The first option, Group data by, specifies the number of Alignments tracks (Figure 2). All will result in only one track, with all the samples on it. Sample creates one track per sample, while Attribute produces one Alignments track per level of the Attribute (i.e. one track per group).
All
Sample
Attribute
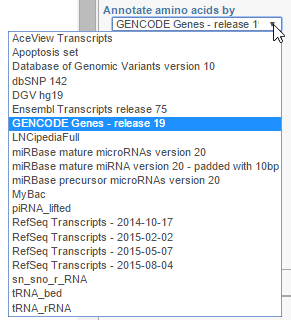
Annotate amino acids by
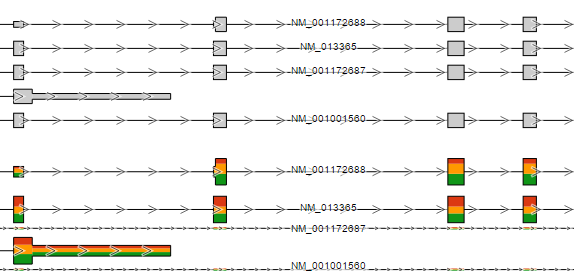
Annotate amino acids by controls the appearance of the Amino acids track and allows you to pick the transcript database that will be used to plot codons (Figure 3). The drop down list shows the databases currently available for the selected genome (additional databases can be added via Library File Management).
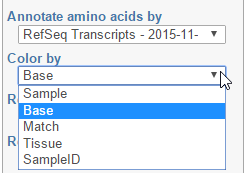
Color by
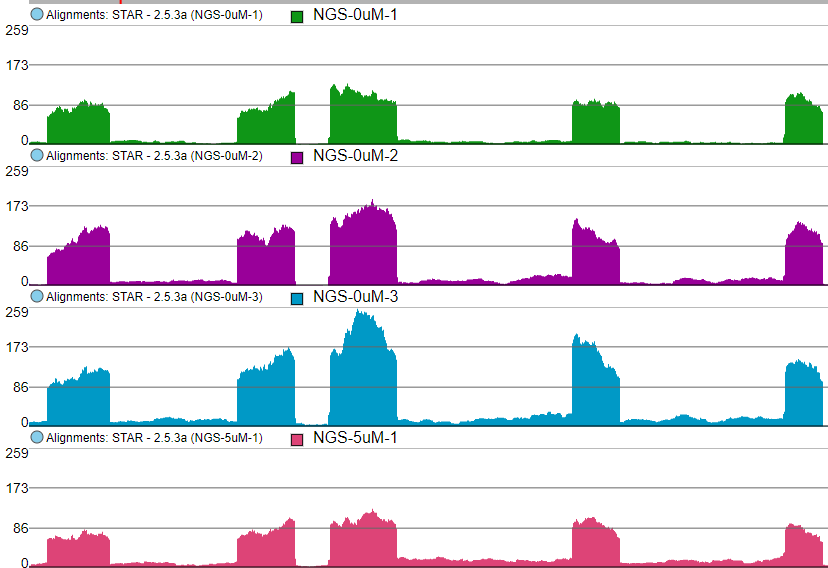
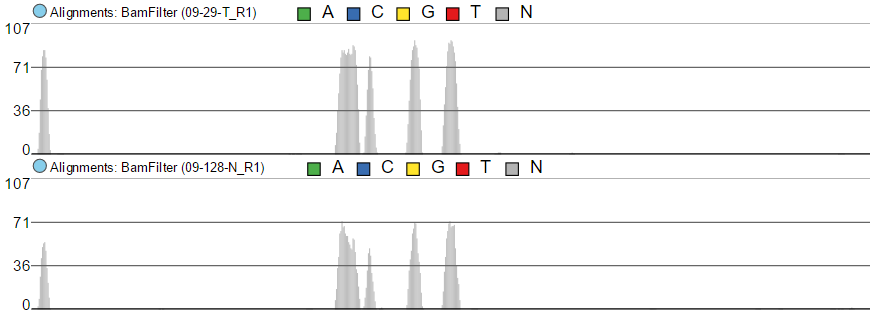
Color by option affects the colouring of the Alignments track and Isoform proportion track. When Sample is selected from the drop-down list, individual samples will be shown on the aforementioned tracks, each sample being given a different colour. If attributes were assigned to samples, they will also be visible in the Color by drop-down (Figure 4) and you will be able to highlight levels of the selected attribute (Figure 5).
Color by Sample
Color by <Attribute>
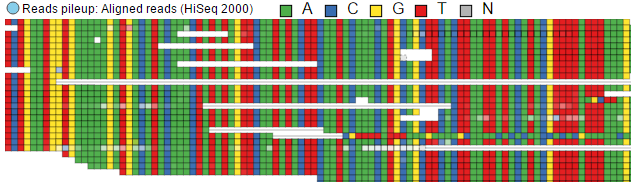
The effect of the option to Color by Base can be seen with high power magnification (Figure 6). Individual base calls are highlighted by different colours. When that option is chosen at low power magnification, all the bases are shown in grey.
Finally, Color by Match can be used to quickly identify mismatches against the reference genome. A matching base is coloured in blue, while mismatch bases are shown in yellow.
Read histogram Y-axis max
The maximum of the y-axis of Alignments tracks is set by Read histogram Y axis scales option (Figure 7). When using Project max, the y-axis for each track is set individually, based on the maximum within that sample. On the other hand, Track max uses the maximum across all the samples and uses that value as the maximum for all.
Project max
Track max
Read histogram type
Read histogram type changes the presentation of the Alignments track and should be used in conjunction with the Group data by and Color by tracks to get the desired visualisation.
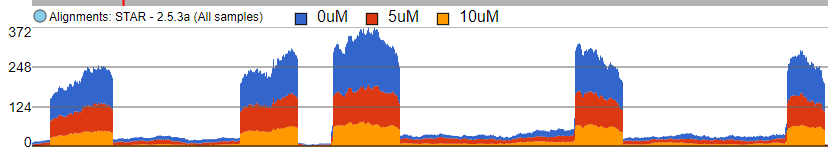
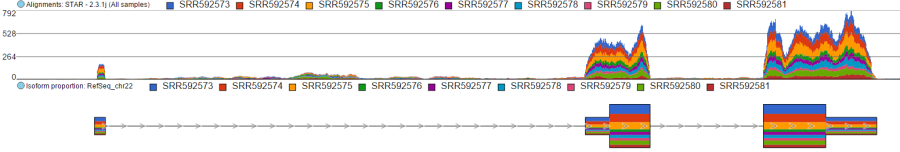
When set to Sum, the Read histogram type shows the sum of base calls at each position, i.e. total coverage per position. Figure 8 shows an Alignments track with three samples. With the Sum option, the number of reads at each base in each sample is added and displayed. The contribution of individual samples is not visible since the track is Colored by Group (but that would make sense in this example).
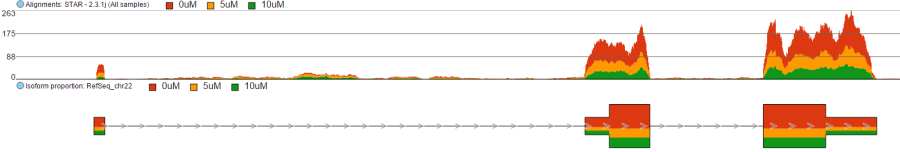
To show the average coverage per locus, switch Read histogram type to Average and leave Color by as is (i.e. by group) (Figure 9). With this setting, Chromosome view will calculate the average by dividing the total coverage per locus by the number of samples. Note that using Color by Sample would not make sense here. Although Figure 8 looks quite like Figure 7, the y-axis range is different.
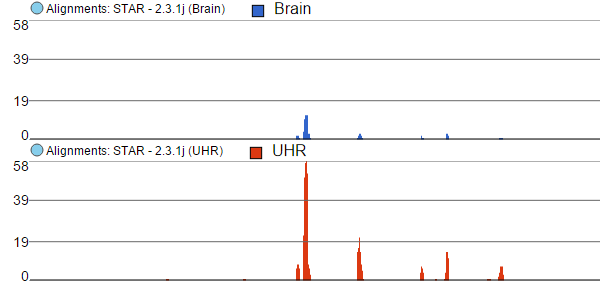
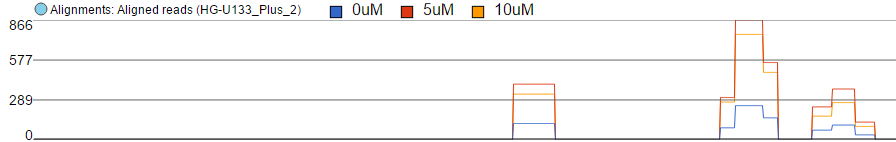
Finally, the option Overlay is useful if you want to directly compare base counts over several samples (or groups) as each will be represented by a line (i.e. no stacking). The example in Figure 10 is based on microarray data, showing three groups on the same Alignments track. The red group has the highest base counts, while the counts in the blue group are much lower.
Split read histogram by strand
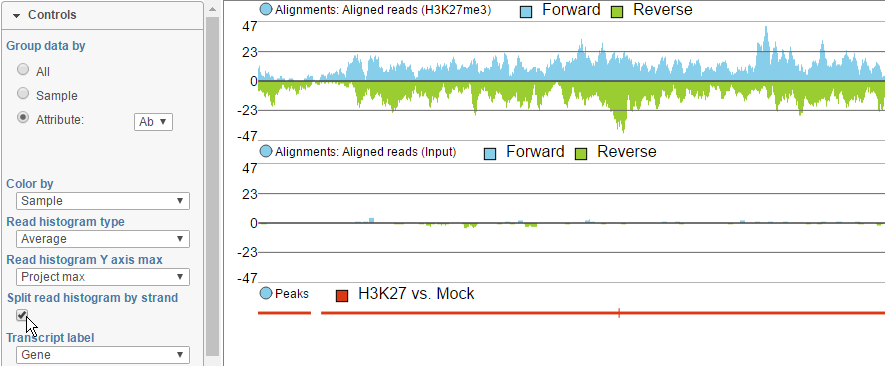
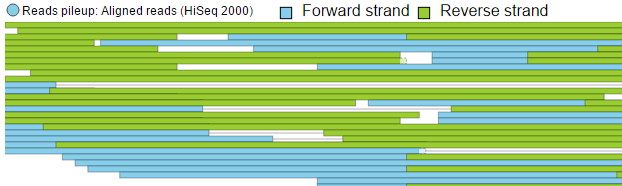
To view the reads histogram grouped by the specific strand that they've mapped to, click the Split read histogram by strand checkbox. This displays the forward reads at the top half of the track and the reverse reads on the bottom half of the track (Figure 11). This track can be helpful in studies such as ChIP-Seq, where strand-specific read distributions can display hallmarks of DNA-protein interactions.
Transcript label
You can use the Transcript label selector to specify labels on the reference transcript track and Isoform proportion track (Figure 12).
Transcript label: Gene
Transcript label: Transcript
Reads pileup and probe color
Reads pileup color: Strand
Reads pileup color: Base
Probe color control customizes the appearance of Probe intensities track (Figure 14). When set to Intensity, the color of a probe reflects its intensity, using a color gradient from white (low) to admiral (high). Alternatively, when Strand is turned on, probes on the reverse strand are in parakeet green, while probe on the forward strand is in sky blue.
Probe color: Intensity
Probe color: Strand
If a variant database is available for the current genome, the variants can be added to the Reference genome track. To show the variants, point the Variant database control to the database of your choice.
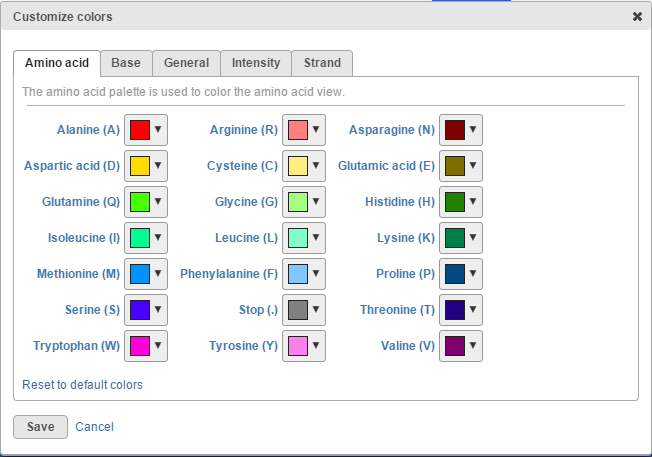
To change any of the colors on the canvas, use the Customize track colors tool. A resulting dialog will help you to pick another color (drop-down button opens the color-picker) (Figure 15).
Track Order
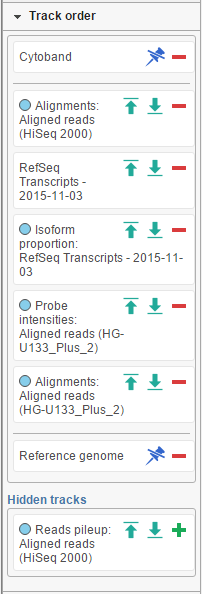
The position of the tracks on canvas can be controlled by using the Track order tool. If you want a track to be visible all the time, i.e. while scrolling up or down, pin it to the top or to the bottom. Below shows the Cytoband track pinnned to the top of the canvas and Reference genome track pinned to the bottom of the canvas. To unpin a track, click on the pin icon ( ). The track will be unpinned and a message No tracks are pinnned to the top / bottom will appear. To pin a track, drag the track name to the No tracks… message. Alternatively, you can use the green arrows ( ) to pin a track. When you mouse over an arrow, the new position of the track will be highlighted on the canvas; click on the arrow to accept.
A track can be hidden (meaning it will not be visible) by selecting the red minus, or unhidden by selecting the green plus icon.
The tracks can be reordered by drag and drop.
Selection Details
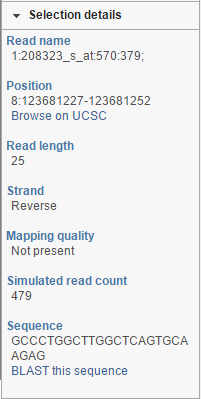
At the bottom of the control panel you will find the Selection details section (Figure 17). It is used to display information on the element selected on the canvas (using the Pointer mode).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
35 | rates |



1 Comment
Melissa del Rosario
author: ilukic