In this section, we will create a list of peaks significantly enriched in the ChIP sample versus the control sample.
- Select Create a list of enriched regions from the Peak Analysis section of the ChIP-Seq workflow
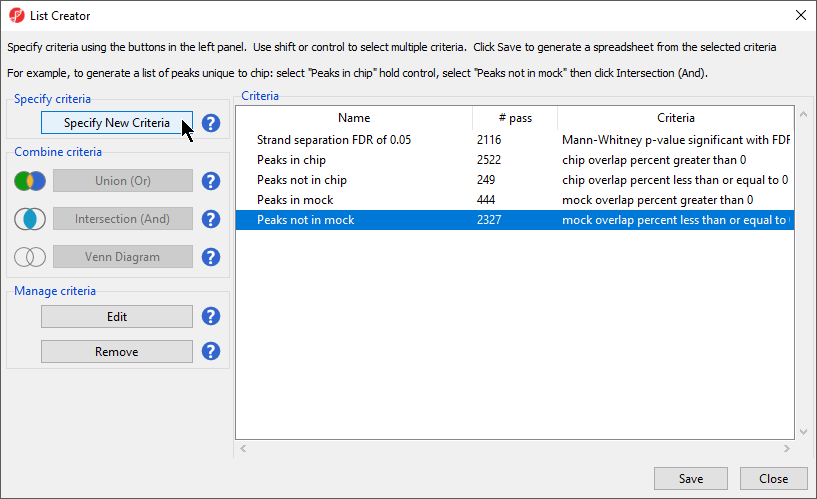
- Select Specify New Criteria (Figure 1)
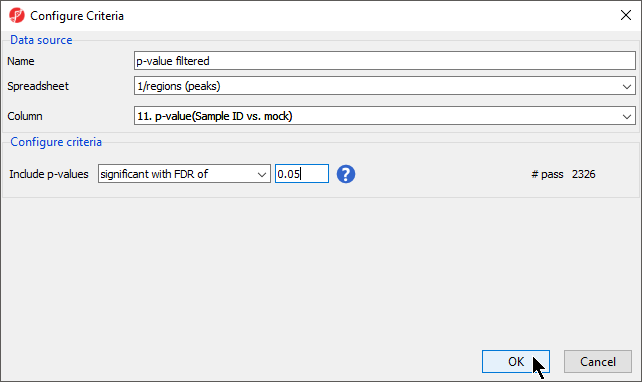
Configure the new criteria as shown (Figure 2).
- Name the criteria p-value filtered
- Select 1/regions (peaks) from the Spreadsheet drop-down menu
- Select 11. p-value(Sample ID vs. mock) from the Column drop-down menu
- Select significant with FDR of from the include p-values drop-down menu with a value of 0.05
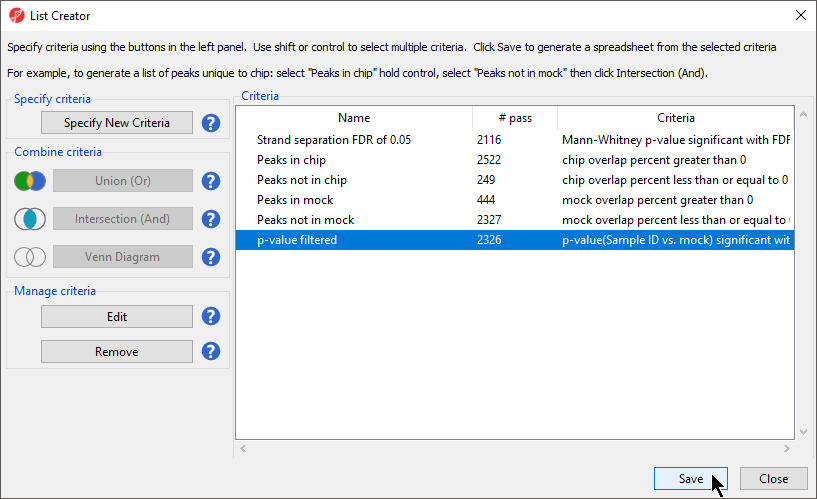
- Select OK to add the criteria to the criteria list (Figure 3)
- Select Save
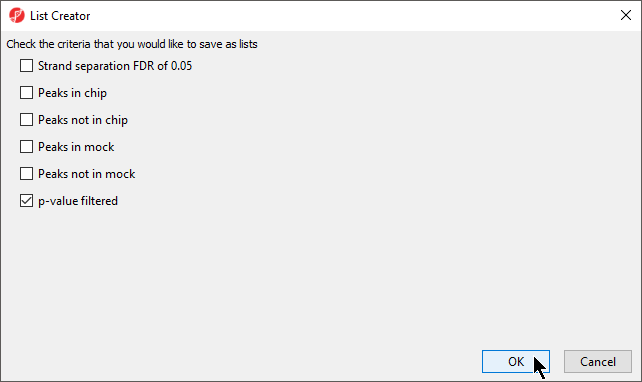
- Select p-value filtered from the list of criteria (Figure 4)
- Select OK
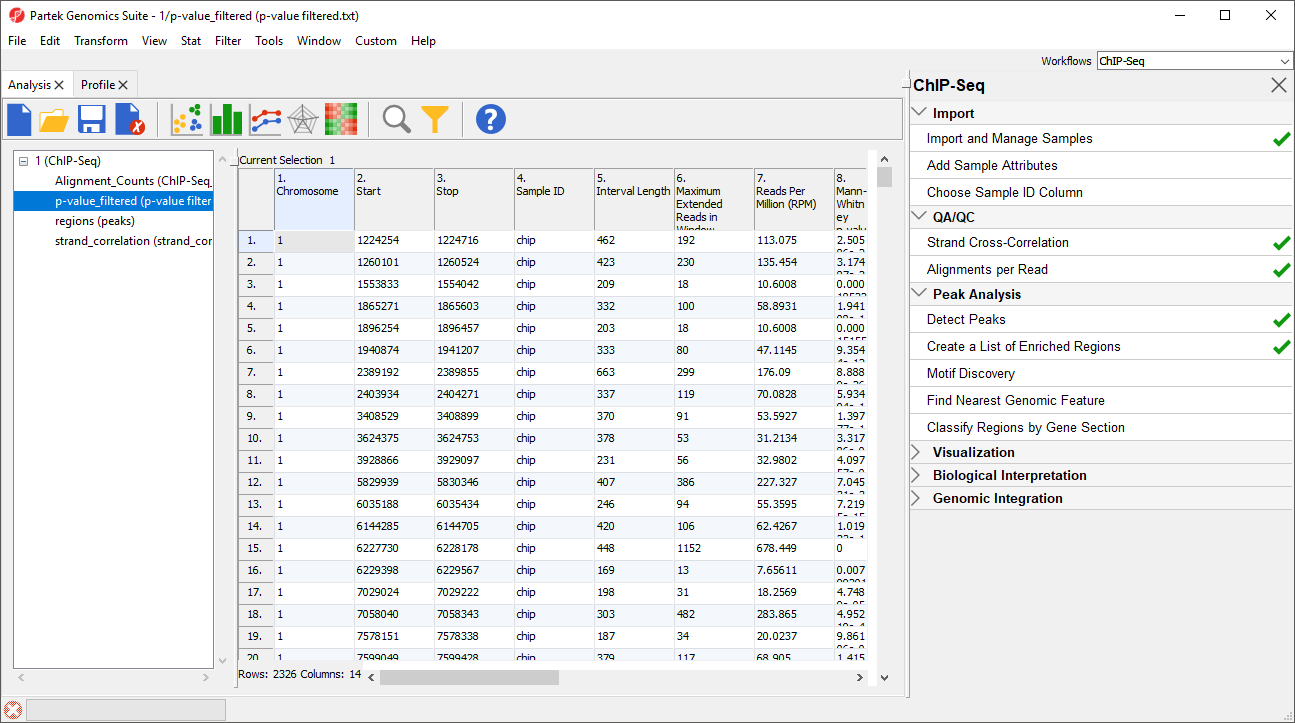
The new spreadsheet will open (Figure 5).
Other List Creator operations like the Venn Diagram, Union (Or), and Intersection (And) of the lists could be used to create different lists of enriched peaks. For example, you could filter on the intersection between Strand Separation FDR of 0.05 and Peaks not in mock or filter by scaled fold change or apply a minimum number of reads per million. The choice of what peaks you want to consider for downstream analysis depends on the goals and details of your experimental design.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
35 | rates |