The setup dialog for GO ANOVA can be found in the Biological Interpretation section of the expression workflows (Gene Expression, MicroRNA Expression, Exon, RNA-Seq, miRNA-Seq). It is recommended that GO ANOVA is run on the sheet with expression levels, after import and normalization, though GO ANOVA can be run on any spreadsheet with samples on rows and genes on columns. If a child spreadsheet is selected, such as the result of a prior ANOVA analysis, then the test will be automatically run on the parent spreadsheet.
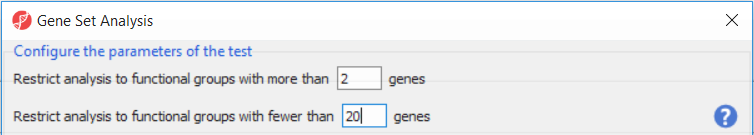
Upon selecting GO ANOVA (Biological Interpretation > Gene Set Analysis), Partek Genomics Suite will first offer the opportunity to configure the parameters of the test and exclude functional groups with too few or too many genes (Figure 1). To save time when running GO ANOVA, the size of GO categories analyzed can be limited using the Restrict analysis to function groups with fewer than __ genes. Large GO categories may be less interesting and also take the most time to analyze. We recommend to restrict the analysis to the groups with fewer than 150 genes, as it can make the analysis much quicker (and the results easier to interpret). In the current example, the maximum category was set to only 20 genes, for demonstration purposes only.
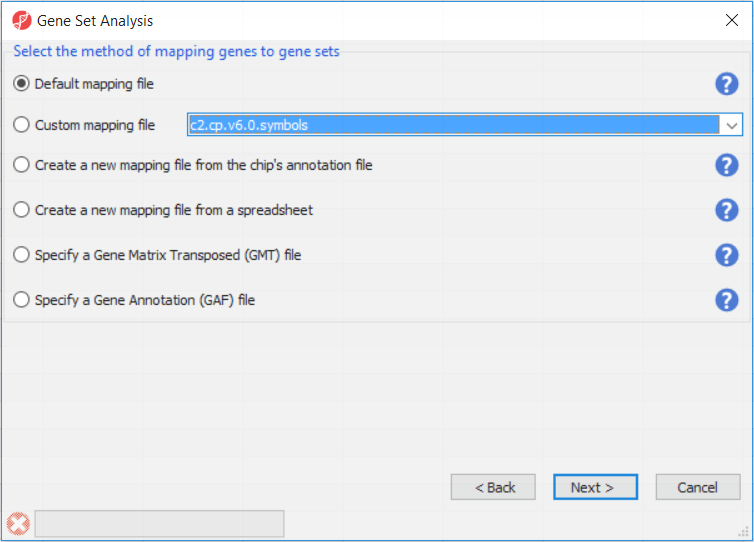
The next dialog (Figure 2) specifies the method of mapping genes to gene sets. Default mapping file is built from annotation files from geneontology.org. Custom mapping file points to the mapping files available on the local computer and present in the Microarray libraries directory. Create a new mapping file from the chip's annotation file option will try to build the annotation file from the annotation file created by the microarray vendor. Create a new mapping file from a spreadsheet enables you to create a custom mapping file from an open spreadsheet, which has gene symbols on one column, and gene groups on the other column. Finally, files in gene matrix transposed (GMT) or gene annotation (GA) formats can also be used.
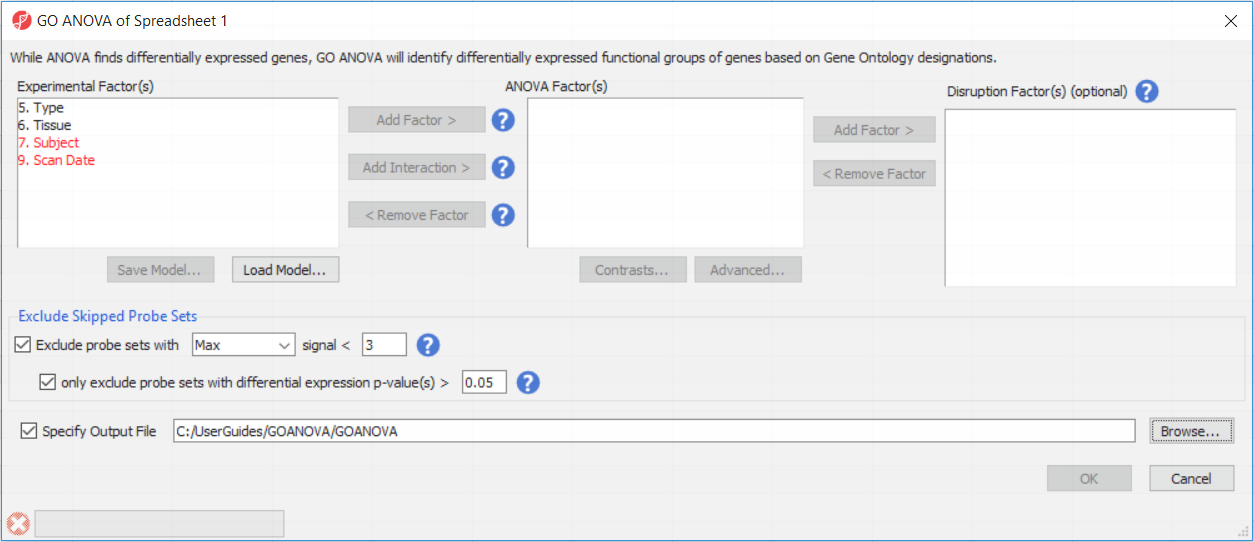
To setup the GO ANOVA dialogue you must consider all factors that would normally be included in an ANOVA model analyzing gene expression among the samples (Figure 3). Briefly this should include:
- Experimental factors
- Factors explaining sample dependence
- Factors explaining noise
Experimental Factors
Factors inherent to the experiment include variables that would be considered as the experimental variables during experiment design. Generally this will include all variables necessary to answer the questions of the researcher. Examples may include factors such as tissue type, disease state, treatment, or dosage.
Sometimes factors do not act independently of each other. For example, different dosages of a drug may affect patients differently over time, or a drug may not affect tissues equally as in many toxicity studies. If the effect of one variable on the other is either suspected of occurring, or of particular interest, an interaction between the two factors should be included. To do this, select the two factors simultaneously by CTRL-clicking the factors and then select Add Interaction.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |