
Variable (V), Diversity (D), and Joining (J) recombination analysis
V(D)J recombination occurs in lymphocytes when T and B cells assemble variable (V), diversity (D), and joining (J) gene segments, contributing to the generation of receptors which recognize and respond to perturbations. V(D)J recombination produces clones of unique T cell receptor (TCR) chains or B cell receptor (BCR) chains giving rise to the diverse repertoire of T and B cell populations which are imperative to adaptive immune system function1. The frequency of generated clones can be measured and explored, giving researchers a powerful view into variation, expansion, and diversity within the biological system. You can import filtered Contig Annotation CSV files2 from the 10X Genomics Cell Ranger V(D)J or multi pipeline3. If there is matching gene expression data, it can also be imported and analyzed within the same project. We recommend uploading the filtered feature barcode matrices as either the Hierarchical Data Format (H5 or HDF5)4 or Market Exchange Format (MEX)5.
Terminology
- UMI (Unique Molecular Identifier): random 10 bp nucleotide sequence that distinguishes which reads came from the same transcript.
- Barcode: the unique identifier in each droplet which usually contains reads from a single cell.
- Contig: assembled reads containing the same UMI and barcode that align to the same transcript sequence.
- Complementarity-Determining Region: CDR1, CDR2, and CDR3 are important in antigen binding of a T or B cell receptor.
- CDR3 (Complementarity-Determining Region 3): CDR3 spans the V(D)J junction. There is one CDR3 nucleotide sequence for each V(D)J contig.
- Clonotype: cells derived from a common ancestor during clonopoiesis which have a particular composition. The cells in a clonotype can have a different number of chains or different CDR3 regions but still be considered a single clone (CDR3 is a highly variable region used for binding; an example of different CDR3 regions would be from affinity maturation which can occur in memory B cells).
Understanding Clone Composition
Multiple cells can have the same clonotype and each clonotype can have multiple makeups. Each clonotype contains one or more chains (TRA and TRB for T cells and IGH, IGK and IGL for B cells), the highest scoring V, D, and J gene segments, and CDR3 nucleotide sequence. T cells have a TRA and TRB chain with V, D, J and C regions. In B cells, IGH is the heavy chain which has a V, D, and J region while IGK and IGL are the light chains with a V and J region. The Immunoglobulins have two identical heavy chains and two identical light chains. B cell isotypes are antigenic determinants that characterize the classes and subclasses of heavy chains and types and subtypes of light chains; the constant region (C gene) produced by the B cell changes but the V regions and specificity do not. Constant regions do not participate in antigen recognition, instead C regions interact to mediate biological function; so isotypes have different function but can bind the same antigen.
Import Data
- Create a new project to upload your data. Ensure that you have transferred the filtered contig_annotations.csv file(s)2 for each sample from either the cellranger vdj6 or cellranger multi7 pipeline to the server, as well as the filtered feature barcode matrices in H5 or MEX format from the cellranger multi pipeline for each sample if you have matching gene expression data.
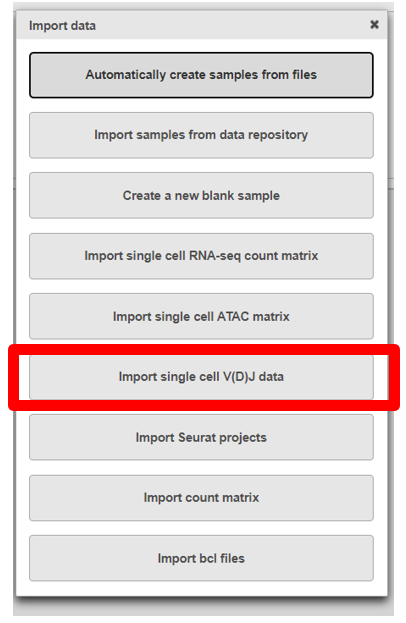
- Click Import, then select Import single cell V(D)J data
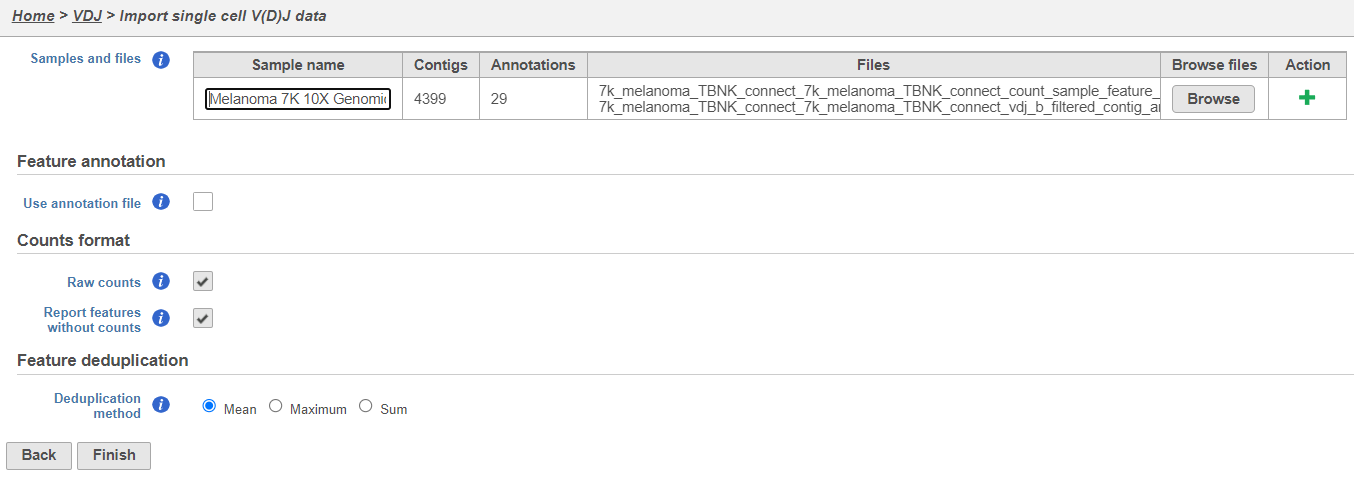
- Upload either the filtered contig_annotations.csv file alone or, if you have matching gene expression, with the filtered count matrix per sample and give each sample a name. To add a sample use the the
 Action. In the example below using default settings, there is one sample with two files, one for V(D)J and one for gene expression. Click Finish.
Action. In the example below using default settings, there is one sample with two files, one for V(D)J and one for gene expression. Click Finish.
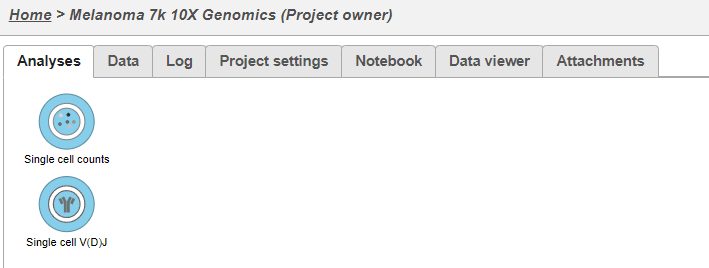
- This results in two starting nodes, one for single cell counts and one for single cell V(D)J as shown below. Note that once subsequent tasks are performed on a node, no more data can be imported into this project. The single cell counts node can be processed as usual; for help information related to this please see the tutorial for Analyzing Single Cell RNA-Seq Data.
References
- Tonegawa, S. Somatic generation of antibody diversity. Nature 302,575–581 (1983). https://doi.org/10.1038/302575a0
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/output/annotation#contig-annotation
- https://support.10xgenomics.com/single-cell-vdj/software/overview/welcome
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/7.0/advanced/h5_matrices
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/7.0/output/matrices
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/using/vdj
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/using/multi


