Peak calling task is used to detect enriched genomic regions on reads generated from ChIP-seq, DNase-seq, MeDIP-seq etc. experiments. Partek® Flow® provides a widely used method MACS2--model-based analysis1 (http://liulab.dfci.harvard.edu/MACS/) to find peaks , it can be used with or without control sample.
MACS2 dialog
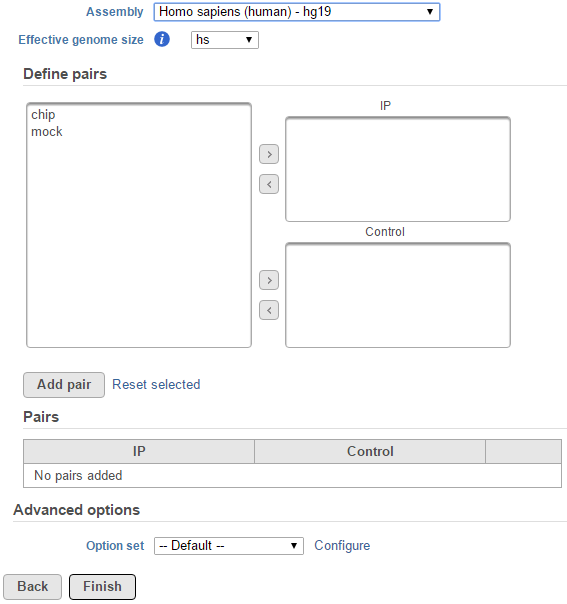
Selecting MACS2 from the context sensitive menu will bring up the MACS2 task dialog, interface will be different depends whether there are sample attributes available in the data tab. If there are only two sample for instance, ChIP and mock as sample name, there is no attribute assigned to the two samples, the dialog will look like (Figure 1).
In the Define pairs section, the left panel list all the sample names, add one pair at a time, select ChIP sample to put in IP panel on the top-right, choose control sample to put in the Control panel on the bottom-right. If there is no control sample in the experiment, the Control panel can be blank. If more than one ChIP or Control samples added, the samples will be combined (or pooled) in the analysis.
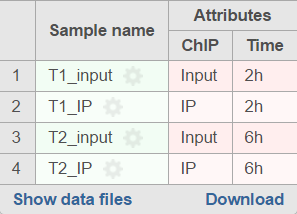
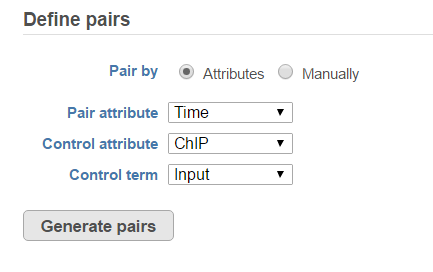
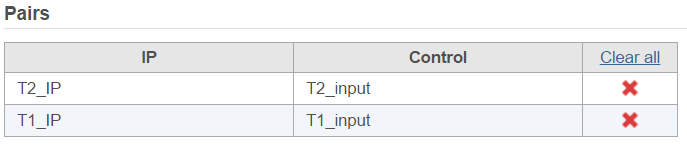
If the sample attributes are defined, you will have an additional option to add pairs based on the attribute. For instance Figure 2 is show an example data with 4 samples, 2 time point, there in one ChIP sample and one input sample in each time point.
References
- Zhang Y, Liu T, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
1 | rates |