What is Space Ranger?
Space Ranger is a set of analysis pipelines that process Visium Spatial Gene Expression data with brightfield and fluorescence microscope images (1).
Space Ranger in Partek® Flow®
Space Ranger 1.3.0 has been wrapped in Partek Flow as Space Ranger task. It does not include all the options and uses cases covered by Space Ranger, but takes .fastq and .jpeg/.tiff files as input and performs alignment, filtering, barcode counting, and UMI counting. The output is gene expression count matrix in a .h5 format (both raw and filtered are available for download via Task Details), which then becomes the starting point for downstream analysis in Partek Flow.
Running Space Ranger in Partek Flow
To run Space Ranger in Partek Flow select Unaligned Reads node on the Analysis tab and then select the Space Ranger task in the toolbox (Figure 1).
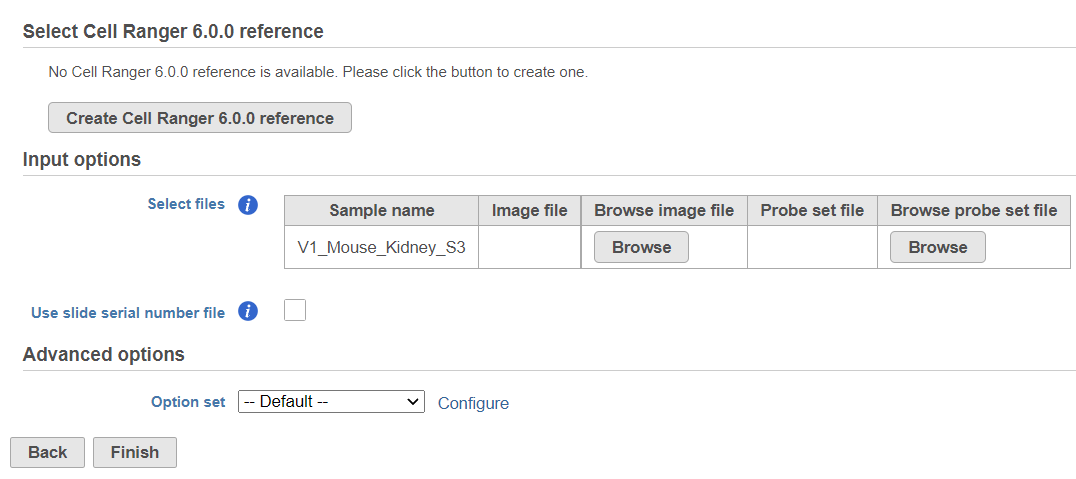
If this is the first time you are using Space Ranger or Cell Ranger, you will need to create a Cell Ranger reference (Figure 2).
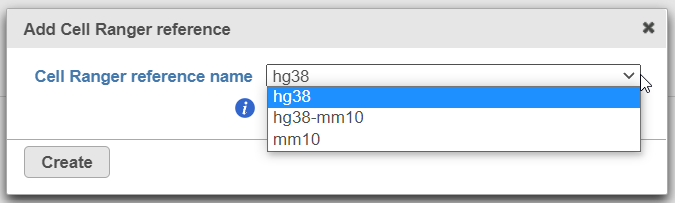
To create a new reference, push the Create Cell Ranger 6.0.0 reference button and pick the appropriate Cell Ranger reference name from the drop down (Figure 3) and then push Create.
Downloading a reference is a separate task and may take some time. However, it will become immediately available in the Cell Ranger 6.0.0. reference drop down (Figure 4).
Alternatively, Cell Ranger reference can be added beforehand, to the Library file management page (on the Other library files tab). At the moment, the following references are available for download from Partek: hg38, mm10, and hg38-mm10 (chimaeric genome).
The sample table under Input options has one row per sample. Image file is required, and that is a single hematoxylin and eosin brightfield image in either .jpg or .tiff format. Click on the Browse button under Browse image file and the file browser will come up. Point to the image file and push Continue. Probe set file is optional; it is a .csv file specifying the probe set used (=target panel).
If you want to specify sample's slide and area information, tick mark the box by the Use slide serial number file and then click Browse to point to the file. The file should be tab-delimited with samples on rows. The first column is the sample name, slide name is on the second column, slide area on the third column.
Click on the Configure link in the Advanced options section to open the Advanced options dialog (Figure 5). Use R1 length to hard trim the R1 reads to specified length; use R2 length to hard trim the R2 reads to specified length. Use the Memory limt option to cap the RAM usage (in GB).