Scatter Plot
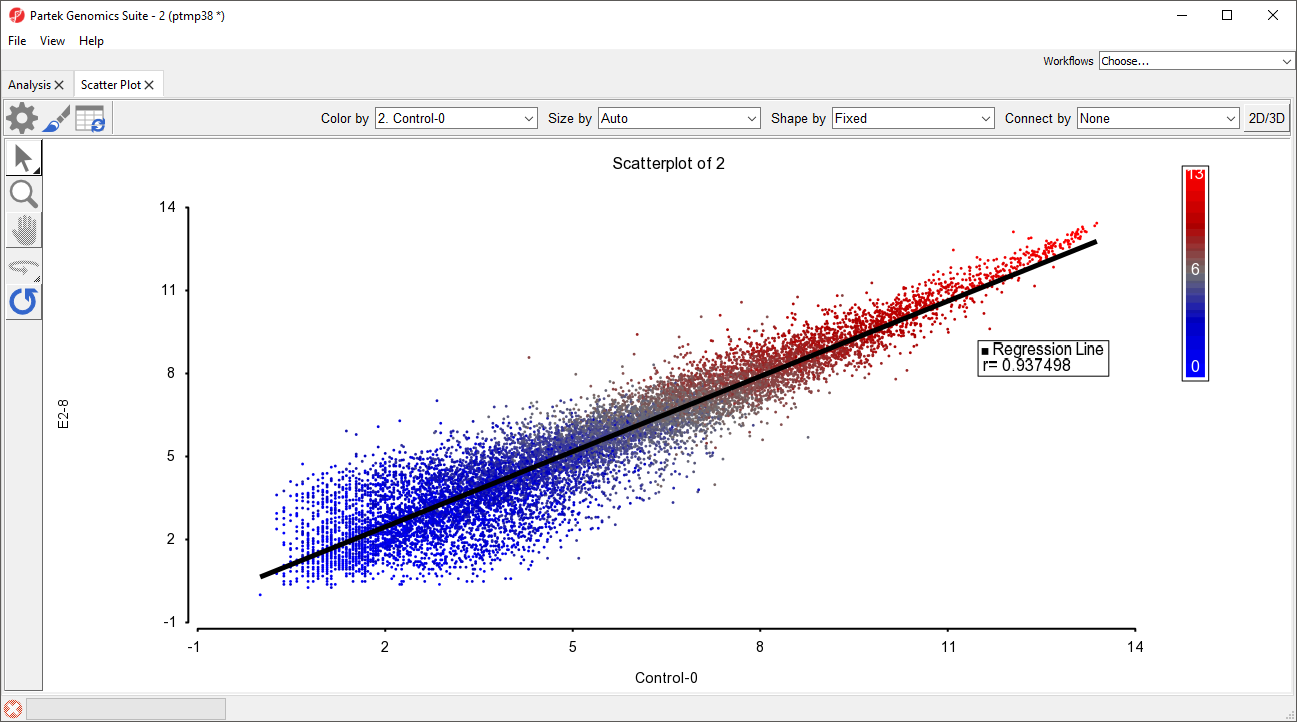
A scatter plot is a simple way to visualize differentially expressed genes. We can plot a scatter plot with gene expression values for two samples at one time. While most probe(sets)/genes fall on a 45° line, up- or down-regulated genes are positioned above or below the line.
To draw a scatter plot, you first need to transpose the original intensities spreadsheet so that the samples are on columns and probe(sets)/genes are on rows.
- Select the main spreadsheet
- Select Transform from the main toolbar
- Select Create Transposed Spreadsheet...
- Select the column with sample IDs from the drop-down menu
- Select OK
A new temporary spreadsheet will be created with probe(sets)/genes on rows and samples on columns.
- Select the two sample columns you would like to compare
- Select View from the main toolbar
- Select Scatter Plot (Figure 1)
- Select Yes when asked if you want to only use the selected columns
- Select Yes when asked if you are sure you would like to draw the scatter plot
The scatter plot will open in a new tab. We can add a regression line to the plot.
- Select () from the plot command bar
- Select Axes
- Select Set Regression Lines
- Select Regression line of y on x
- Set Line Width to 5
- Select OK (Figure 2)
- Select OK to close the Plot Rendering Properties dialog
The scatter plot now features a regression line dividing the probe(sets)/genes (Figure 3).
MA Plot
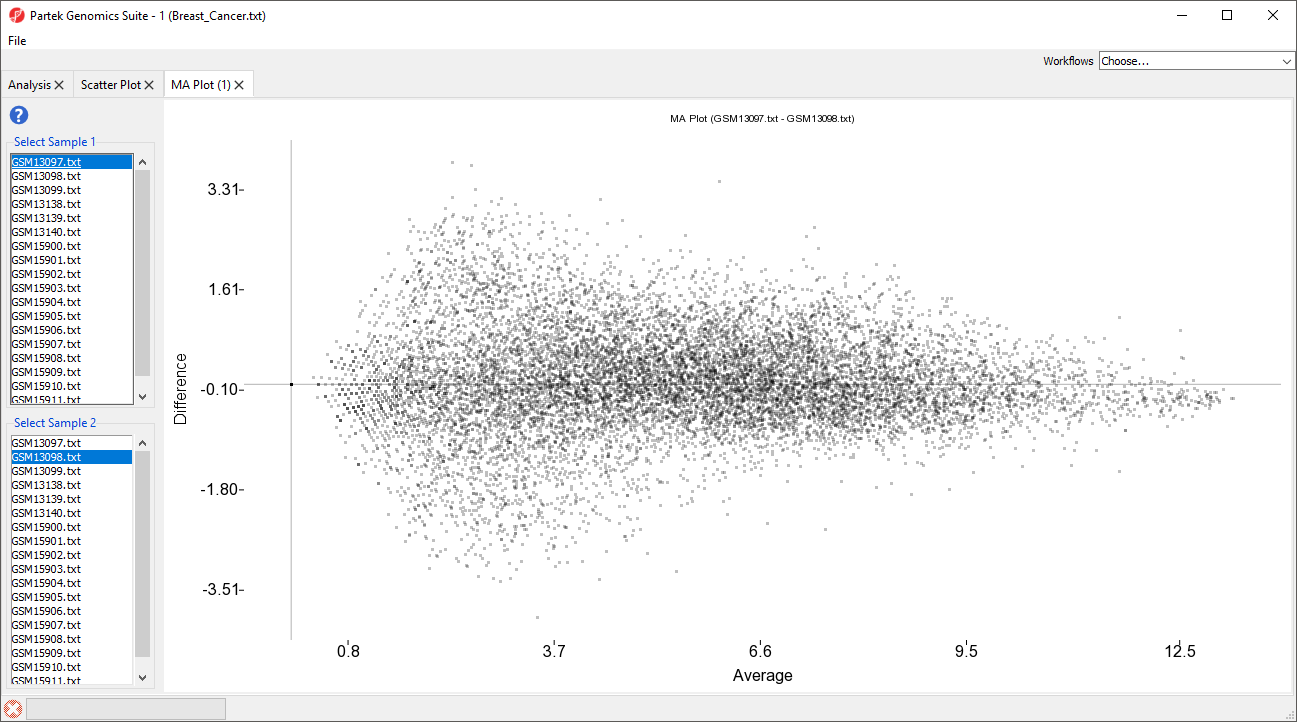
The MA plot can be used to disaply a different in expression patters between two samples. The horizontal axis (A) shows the average intensity while the vertical axis (M) shows the intensity ratio between the two samples for the same data point. In essence, an MA plot is a scatter plot tileted to the side so that the differentially epxressed probe(sets)/genes are located above or below the 0 value of M. An MA plot is also useful to visualize the results of normalization where you would hope to see the median of hte values follow a horizontal line.
The MA plot is invoked on the original intensities spreadsheet with any need for transposition.
- Select View from the main toolbar
- Select MA Plot
The MA plot will launch in a new tab showing the first two rows as the comparison (Figure 4).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
9 | rates |