We will now examine the results of our exploratory analysis and use a combination of techniques to classify different subsets of T and B cells in the MALT sample.
Exploratory Analysis Results
- Double click the UMAP data node
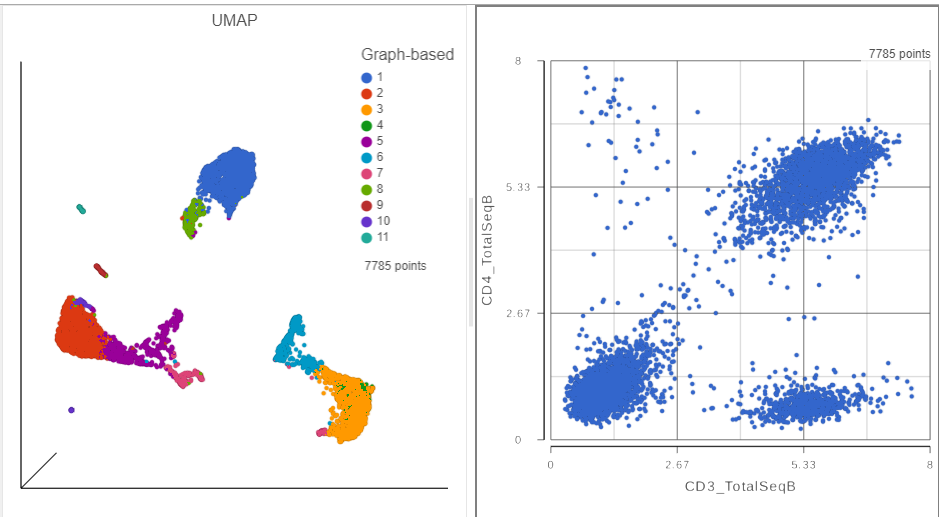
- In the Configuration card on the left, expand the Color card and color the cells by the Graph-based attribute (Figure ?)
- Click and drag the 2D scatter plot icon from the Available plots card onto the canvas (Figure ?)
- Drop the 2D scatter plot to the right of the UMAP plot
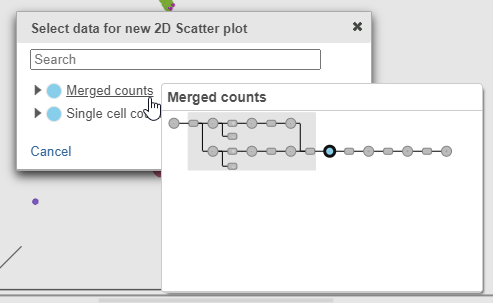
- Click Merged counts to use as data for the 2D scatter plot (Figure ?)
- In the Selection card on the right, click Rule to change the selection mode
- Click the blue circle next to the Add rule drop-down menu (Figure ?)
- Click Merged counts to change the data source for the drop-down list
- Choose CD3_TotalSeqB from the drop-down list (Figure ?)
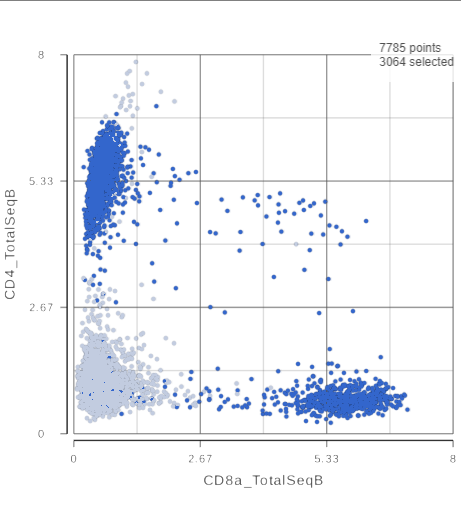
- Click and drag the slider on the CD3D_TotalSeqB selection rule to include the CD3 positive cells (Figure ?)
- Click Merged counts in the Data card on the left
- Click and drag CD8a_TotalSeqB onto the 2D scatter plot (Figure ?)
- Drop CD8_TotalSeqB onto the x-axis option
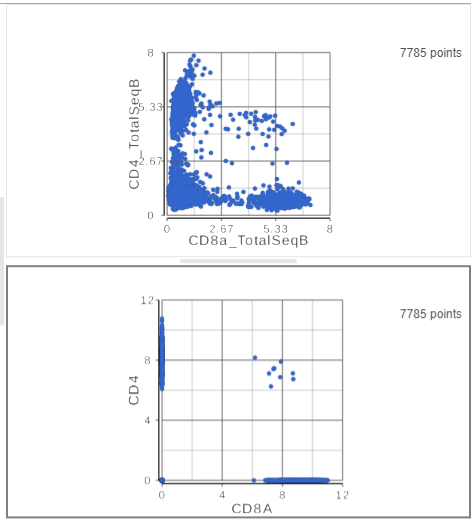
- Click the duplicate plot icon above the 2D scatter plot (Figure ?)
- Click Merged counts in the Data card on the left
- Search for the CD4 gene
- Click and drag CD4 onto the duplicated 2D scatter plot
- Drop the CD4 gene onto the y-axis option
- Search for the CD8A gene
- Click and drag CD8A onto the duplicated 2D scatter plot
- Drop the CD8A gene onto the x-axis option
The second 2D scatter plot has the CD8A and CD4 mRNA markers on the x- and y-axis, respectively (Figure ?).
- On the second 2D scatter plot, click in the top right corner
- Manually select the cells with high expression of the CD4 gene marker (Figure ?)
T cells
B cells
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |