Partek Flow supports .bcl files based on 10X Genomics library preparation. The following document will guide you through the steps.
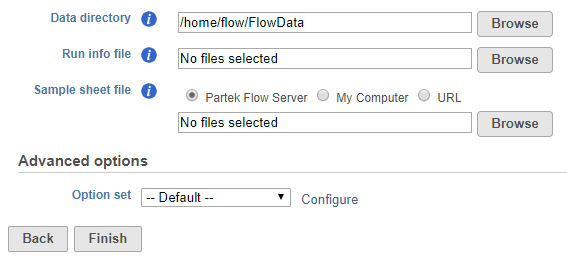
To start the import, create a new project and then select Import Data > Import bcl files. The Import bcl dialog will come up (Figure 1).
Use the Data directory option to point to the location of the directory holding the data. It is located at the top level of the run directory and is typically labeled Data. Please see the tool tip for more info.
Use the Run info file option to point to the RunInfo.xml file. It is located at the top level of the run directory.
Us the Sample sheet file to point to the sample sheet file, which is usually a .csv file.
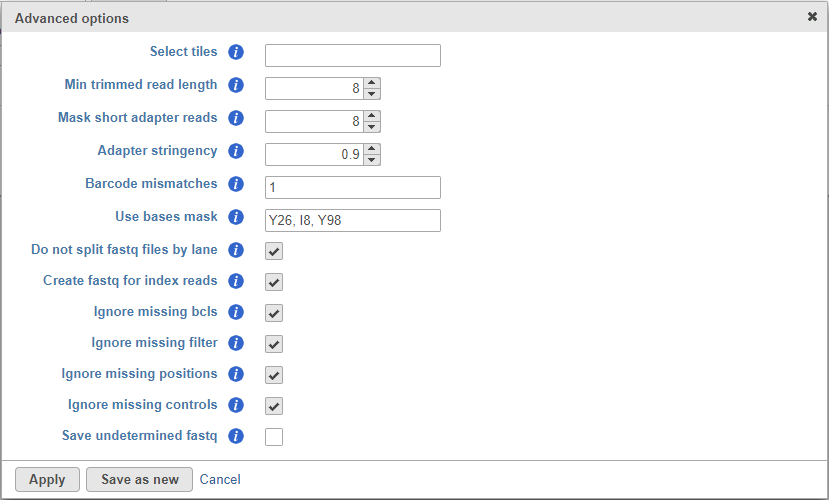
The click on the Configure link and make the following changes (Figure 2).
- Min trimmed read length: 8
- Mask short adapter reads: 8
- Use basis mask: Y26, I8, Y98
- Create fastq for index reads: ON
- Ignore missing bcls: ON
- Ignore missing filter: ON
- Ignore missing positions: ON
- Ignore missing controls: ON
Click Apply to accept and then Finish to import your files.
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
5 | rates |