Partek Genomics Suite enables you to visualize each probe and compare the methylation between the groups at a single CpG site level.
- Right click row 19. Cdk14 in the LCLs vs B cells spreadsheet
- Select Browse to Location from the pop-up menu
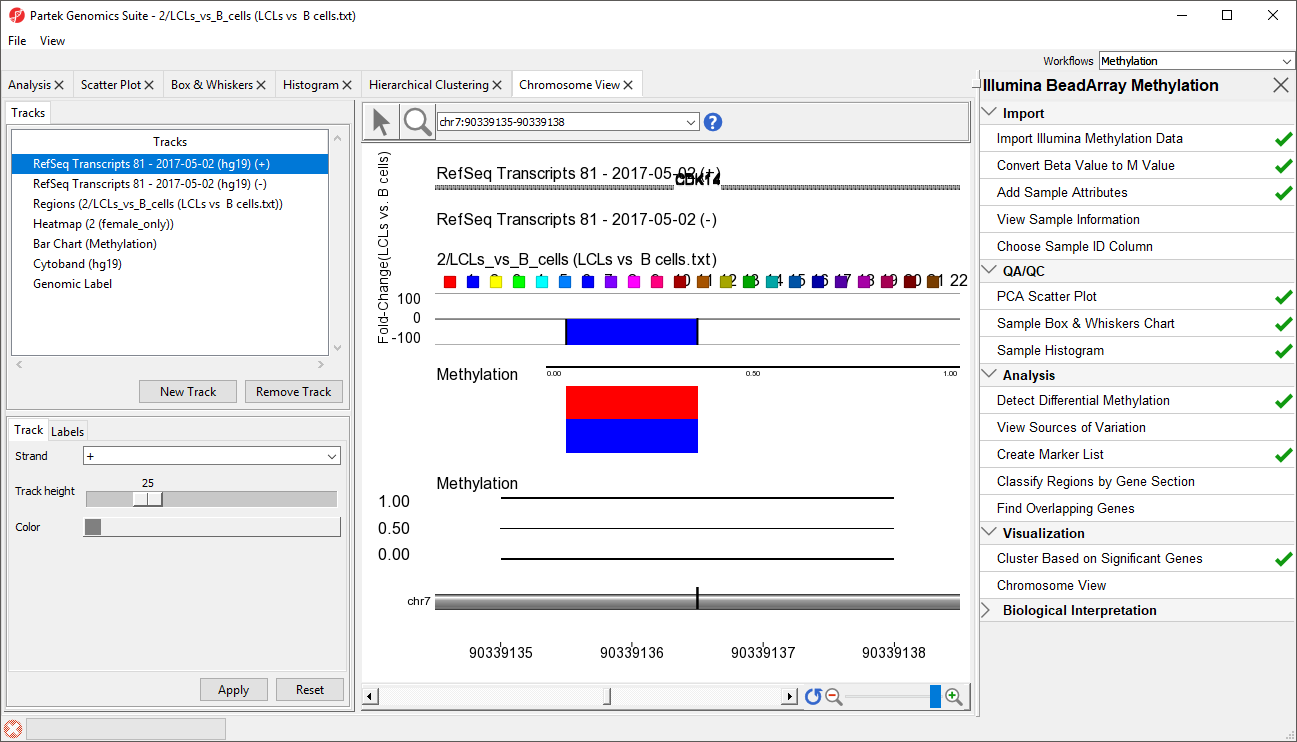
- RefSeq Transcripts 2017-05-02 (hg19) (+): transcripts coded by the positive strand
- RefSeq Transcripts 2017-05-02 (hg19) (-): transcripts coded by the negative strand
- Regions: by default, fold-change difference in methylation between the groups
- Heatmap: M values for all the samples
- Barchart: methylation levels of the selected sample (to select a sample, click on a heat map)
- Cytoband: cytobands of the current chromosome
- Genomic Label: coordinates on the current chromosome
To modify a track, select it in the Tracks panel to bring up its configuration options panel below the Tracks panel. Let's modify a few tracks to improve our visualization of the data.
- Select the Regions track, opens to Profile tab
- Set Bar height by to Estimate
- Set Min and Max to -10 and 10
- Select Apply to change
- Select Color tab
- Set Color bars by to Fold Change (LCLs vs. B cells) (Description)
- Select Apply to change
This will allow us to visualize the differences in methylation between the two groups.
- Select the Bar Chart track, opens to Y-axis tab
- Set Min and Max to -5 and 5
- Select Apply to change
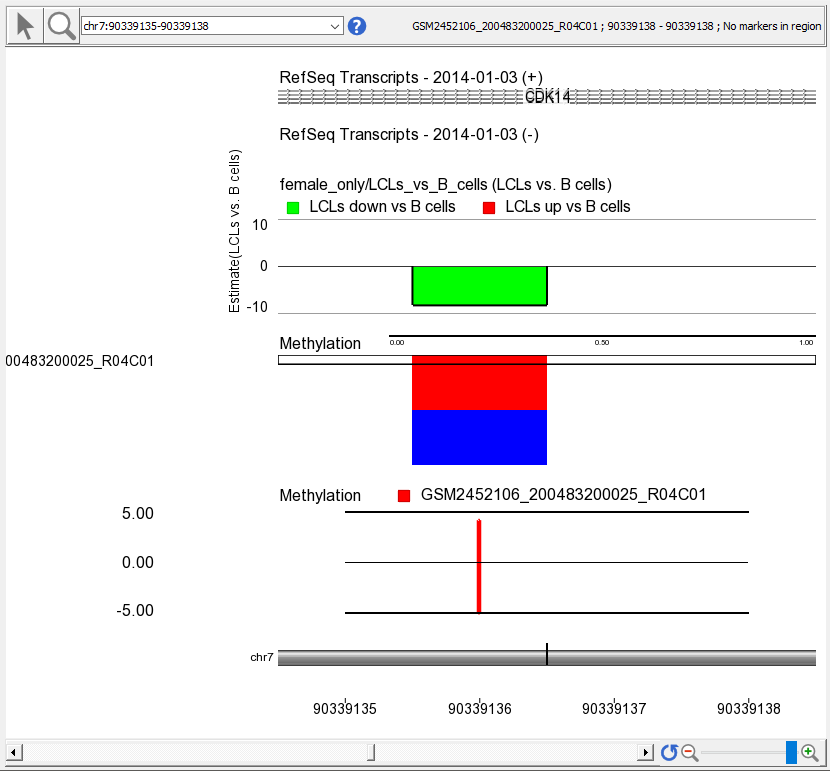
- Select a sample to display in the Bar Chart track by left-clicking on a row in the heat map
A bar indicating the M value at this probe set will appear in the Bar Chart track (Figure 3).
The New Track button allows new tracks to be added to the viewer, while the Remove Track button removes the selected track from the viewer. Tracks can be reordered by selecting a track in the Tracks panel and dragging it up or down to move it in the list. In the Genome Viewer, select () for selection mode and () for navigation mode. In navigation mode, left-click and draw a box on any track to zoom in. All tracks are synced and will zoom together. Zooming can also be controlled using the interface in the lower right-hand corner of the tab (). View can be reset to the whole chromosome level using reset zoom (). Searching for a gene or transcript in the position box will also zoom directly to its location.
The available tracks can be supplemented with a special annotation file that can be built using a UCSC annotation file as the basis. Building and viewing the UCSC annotation file is available as an optional section of the tutorial, Optional: Add UCSC CpG island annotations.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |