There are many useful visualizations, annotations, and biological interpretation tools that can operate on a gene list. In order for these features work with an imported list, an annotation file must be associated with the gene list. Additionally, many operations that work with a list of significant genes (like GO- or Pathway-Enrichment) require comparison against a background of “non-significant” genes. The quickest way to accomplish both is to use the background of “all genes” for that organism provided by an annotation source like RefSeq, Ensembl, etc. in .pannot (Partek® annotation), .gff, .gtf, .bed, tab- or comma-delimited format. If the file is not already in a tab-separated or comma delimited format, you may import, modify, and save the file in the proper file format.
Associating a spreadsheet with an annotation file
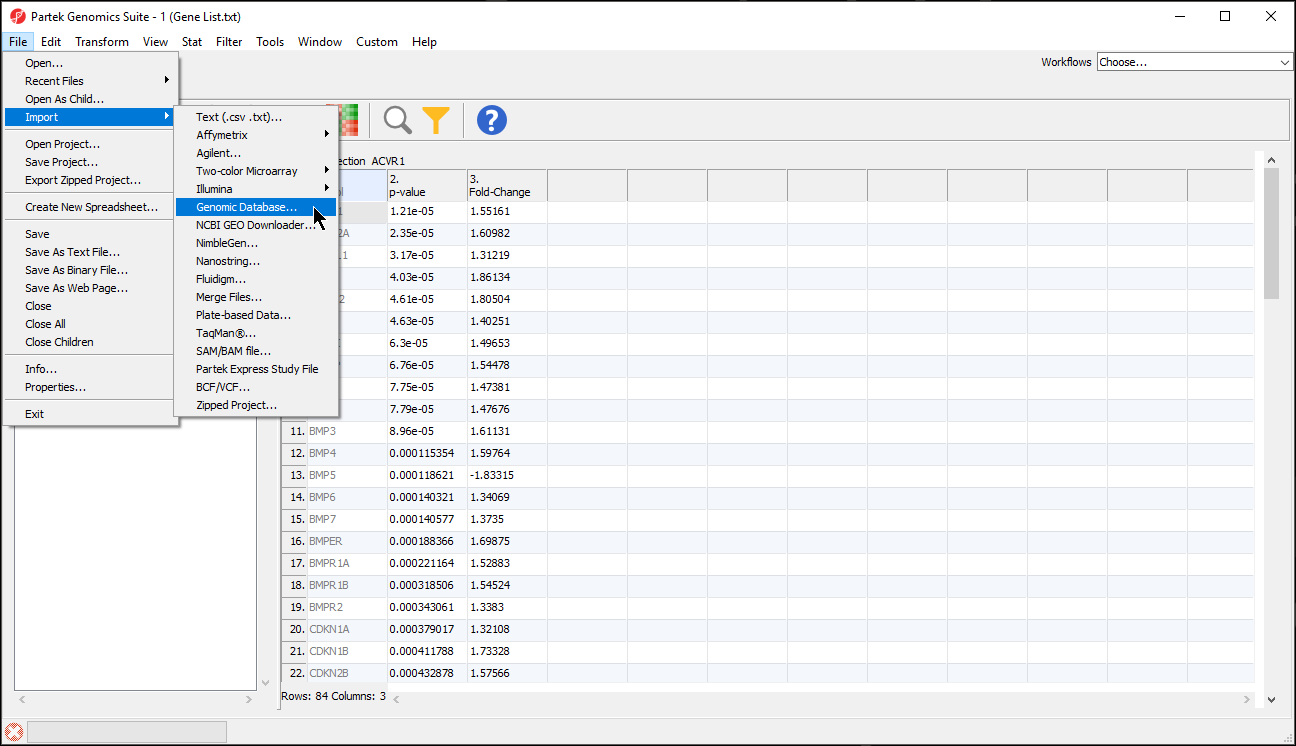
- Select File from the main toolbar
- Select Genomic Database under Import (Figure 1)
- Select the annotation file; we have selected hg19_refseq_14_01_03_v2.pannot from the C:/Microarry Libraries folder
- Delete or rearrange the columns as necessary; we have placed the column with identifiers that correspond to our gene list first
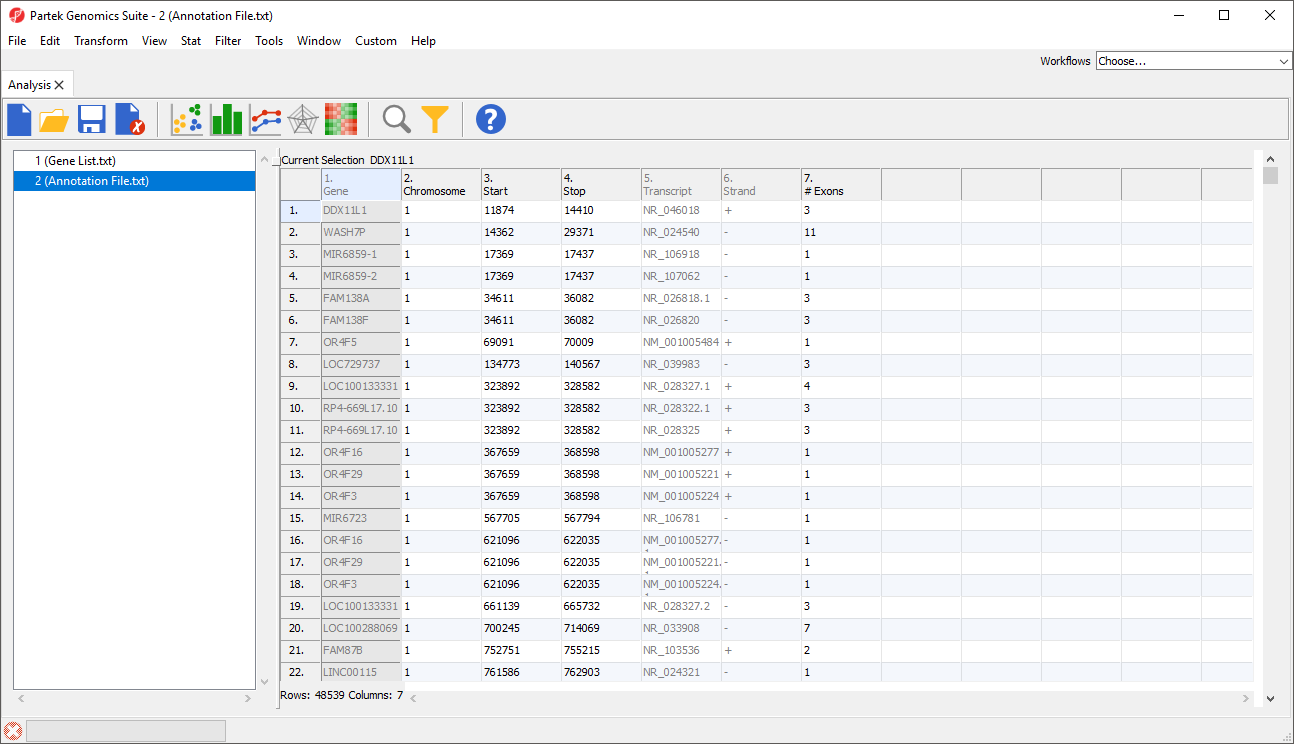
- Select File then Save As Text File... to save the annotation file; we have named it Annotation File (Figure 2)
- Select () to close the annotation file
Now we can add the annotation file to our imported gene list.
- Right click 1 (gene_list.txt) in the spreadsheet tree
- Select Properties from the pop-up menu
This brings up the Configure Genomic Properties dialog (Figure 3).
- Select Browse under Annotation File
- Choose the annotation file; we have chosen Annotation File.txt
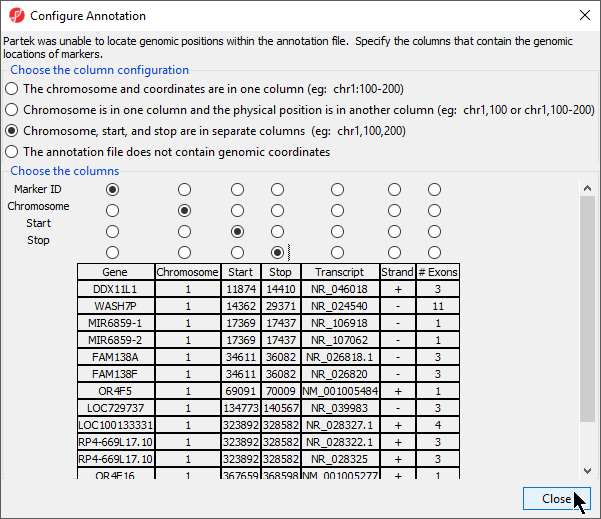
If this is the first time you have used an annotation, the Configure Annotation dialog will launch. This is used to choose the columns with the chromosome number and position information for each feature. Our example annotation file has chromosome, start, and stop in separate columns.
- Select the proper column configuration options (Figure 4)
- Select Close to return to the Configure Genomic Properties dialog
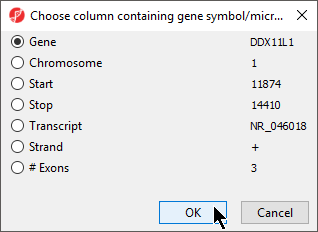
- Select Set Column: to open the Choose column with gene symbols or microRNA names dialog (Figure 5)
- Select the appropriate column; here the default choice of 1. Symbol is appropriate
- Select OK to return to the Configure Genomic Properties dialog
- Select the appropriate species and genome build options; we have selected Homo sapiens and hg19 (Figure 6)
- Select OK
- Select () to save the spreadsheet
The annotation file has been associated with the spreadsheet and additional tasks can now be performed on the data.
Adding annotations to a spreadsheet
Inserting annotations from an annotation file
If an annotation file has been associated with a spreadsheet, annotations from the file can be added as columns in the spreadsheet.
- Right click on a column header
- Select Insert Annotation
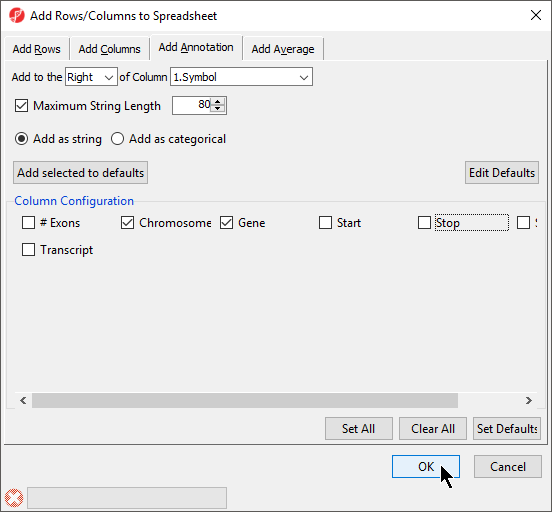
- Select columns to add from Column Configuration; we have selected Chromosome, Start, and Stop (Figure 7)
- Select OK
Annotating with cytobands
- Select Annotate with Cytobands from Tools in the main toolbar when a suitable spreadsheet is open
A column with cytoband locations will be added to the spreadsheet. Adding a cytoband is possible if genomic coordinates are associated with the gene list spreadsheet.
Annotating with known SNPs
- Select Annotate with Known SNPs... from Tools in the main toolbar when a suitable spreadsheet is open
A column of SNPs associated the listed genes and a column indicating the number of SNPs known to be associated with the genes will be added to the spreadsheet. If a SNP database has not been previously downloaded, it will need to be downloaded through the SNP database dialog (Figure 8).
Alternatively, to generate a list of SNP IDs per row, right-click on a row header and select Create list of dbSNP.
In addition to SNPs, this feature can associate any data with a list of genes or genomic coordinates; the dbSNP database, any miRNA database, data from the Database of Genomic Variants (dgv), any mRNA transcriptome database, or any custom annotation source can be associated with your list. In each case, this feature will add columns to the imported gene list spreadsheet that match the genes with features from those databases.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |