With the GO Enrichment feature in Partek Genomics Suite software, you can take a list of significantly expressed genes/transcripts and find GO terms that are significantly enriched within the list. For a detailed introduction of GO Enrichment, refer to the GO Enrichment User Guide (Help > On-line Tutorials > User Guides).
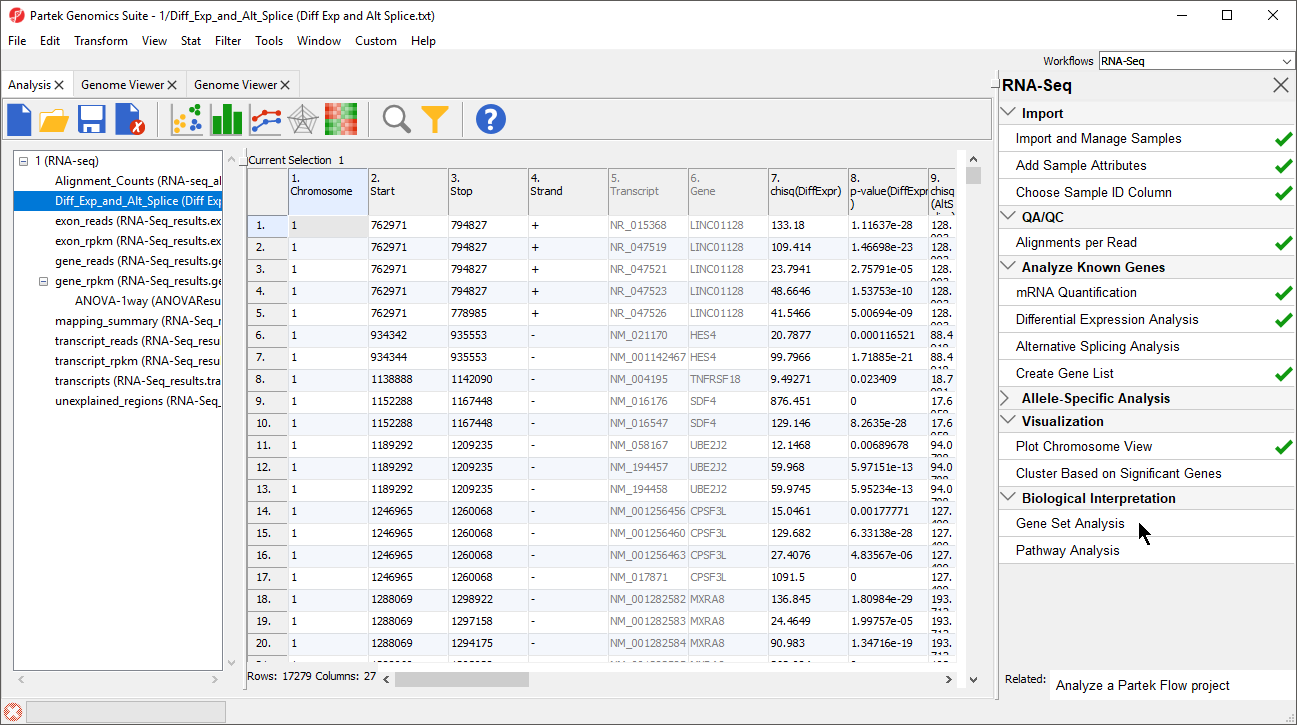
- Select Gene Set Analysis in the Biological Interpretation section of the RNA-seq workflow (Figure 1)
Figure 1. Selecting Gene Set Analysis
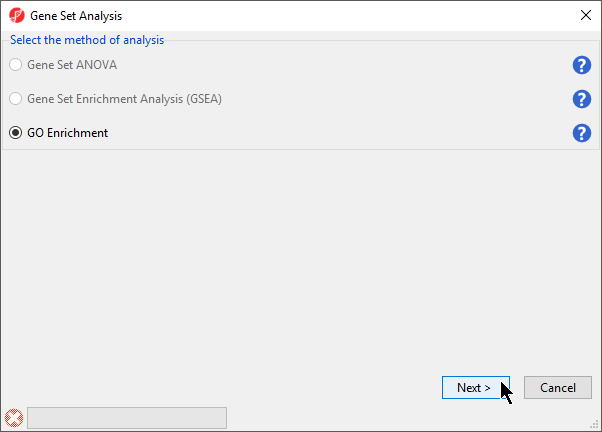
- Select GO Enrichment in the Gene Set Analysis dialog (Figure 2)
- Select Next >
Figure 2. Selecting the method of analysis
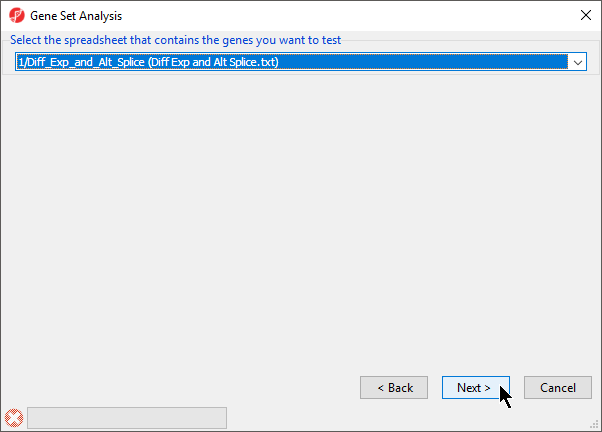
- Select the spreadsheet 1/Diff_Exp_and_Alt_Splice (Diff Exp and Alt Splice.txt) from the drop-down menu (Figure 3)
- Select Next >
Figure 3. Selecting the spreadsheet that contains the genes you want to test
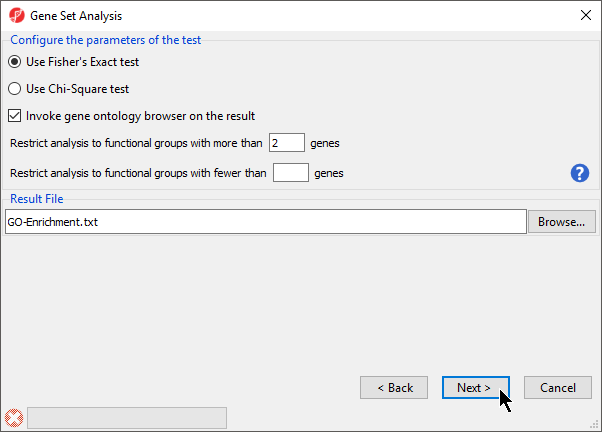
- Select Use Fisher's Exact test
- Select Invoke gene ontology browser on the result
- Set Restrict analysis to functional groups with more than _ genes to 2 (Figure 4)
- Select Next >
Figure 4. GO Enrichment options
- Select Default mapping file (Figure 5)
- Select Next >
Figure 5. Selecting mapping file for GO Enrichment analysis
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |
Overview
Content Tools