Page History
...
Because genomics datasets are generally large, it is ideal to have the data copied in a folder directly accessible to the Partek Flow server. Make sure that the directory has the appropriate permissions for Partek Flow to read and write files in that folder. You may wish to seek assistance from your system administrator in uploading your data directly.
Select the files you would like to create samples from. Once they've been created, assign the corresponding sample attributes for each sample using the Metadata tab. The most efficient way to assign sample attributes is by clicking Assign sample attributes from a file and uploading a tab delimited text file. The file should contain a table with the following:
...
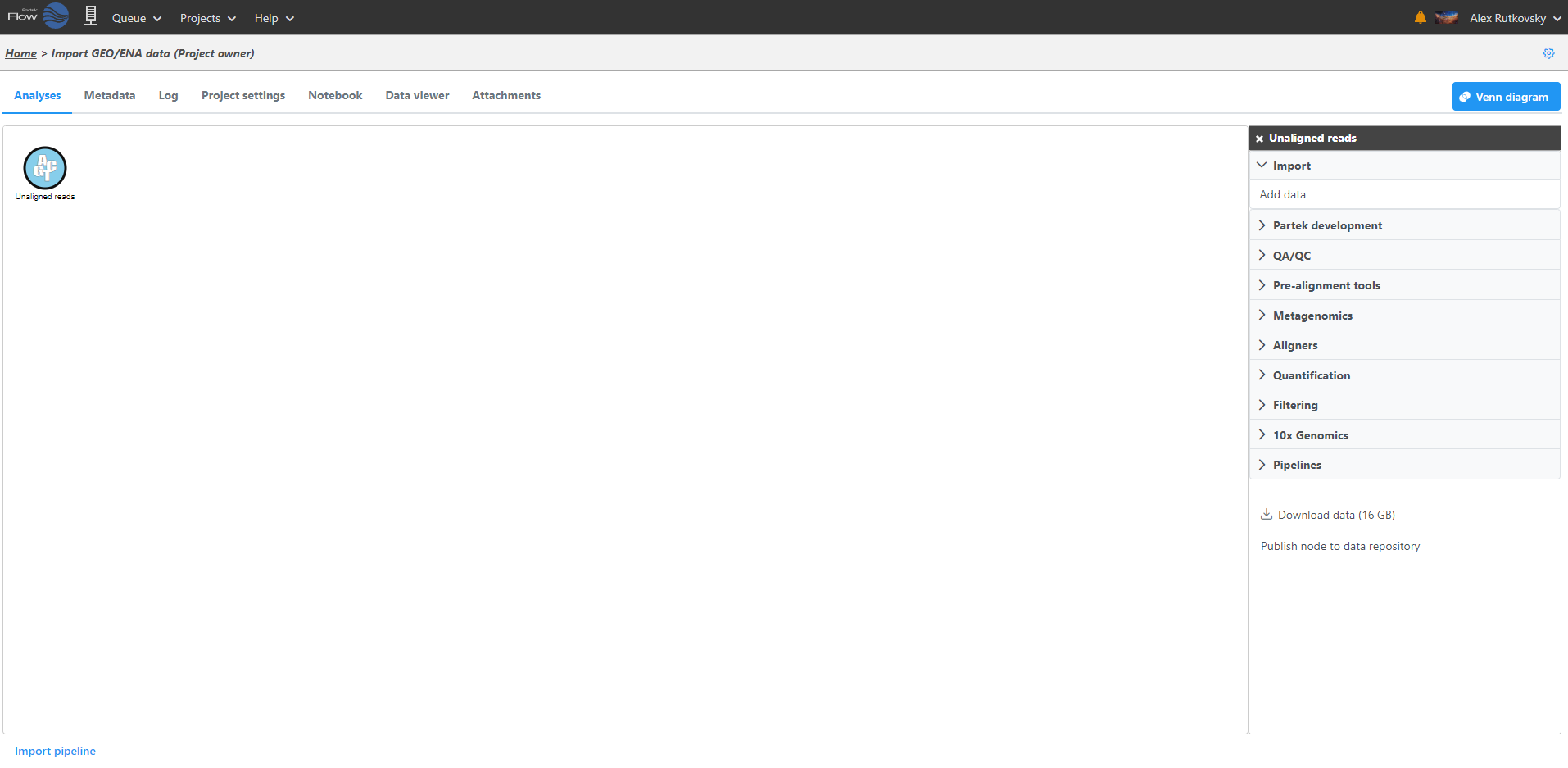
After samples have been added and associated with valid data files, a data node will appear in the Analyses tab (Figure 3). The Analyses tab is where you can invoke tasks, using the context sensitive menu on the right, and view the results of your analysis.
To add more data use the Add data task in the menu on the right or Add data in the Metadata tab. Once a task is performed, data can no longer be added to the project.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...