Page History
...
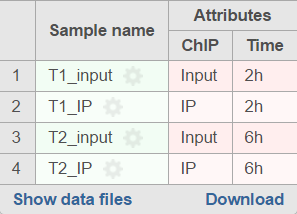
If the sample attributes are defined, you will have an additional option to add pairs based on the attribute. For instance Figure 2 is show an example data with 4 samples, 2 time point, there in one ChIP sample and one input sample in each time point.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
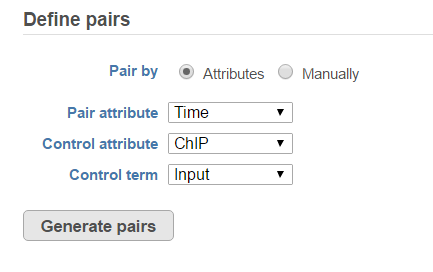
When select MACS2 task, the default option is to use sample attribute to add multiple pairs at one button click (Figure 3)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
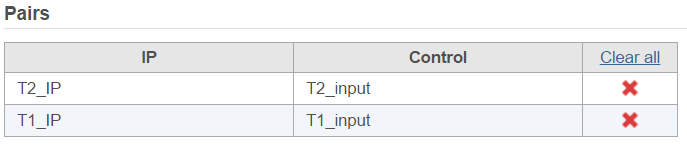
There are IP-Input pair in each time point, so the pair attribute is Time; Control attribute is the attribute contains IP and input group, which is ChIP, the control term is labeled as Input in the example, when click Generate pairs, the two pairs will be automatically added to the Pairs table at once (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Peaks report
The peaks report is generated on each pair specified in the Pairs table separately (Figure 4)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
Absolute summit:
Pileup
-log10(pvalue)
Fold enricment
-log10(qvalue)
References
- Zhang Y, Liu T, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137.
...