Page History
...
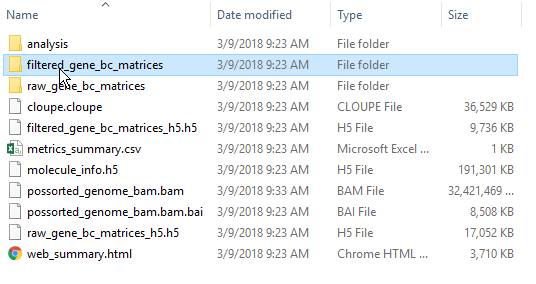
You must first transfer the files to the server. Use the Transfer files button on the Partek® Flow® homepage or use the transfer files link to upload data to the server. Navigate to the folder containing your filtered matrices. By default, the Cell Ranger pipeline output will have a folder called filtered_gene_bc_matrices (Figure 3). It is helpful to rename and organize the files prior to transfer using the File browser. Use drag and drop or click My device to browse files from the device. Click Browse to modify the upload directory. The upload directory should be specified, known, and distinguishable for project file management. You will return to this directory and access the files to import them into a project.
...
...
By default, the Cell Ranger pipeline
...
output will have a folder called filtered_gene_bc_matrices
...
(Figure 3). It is helpful to rename and organize the files prior to transfer using the File browser.
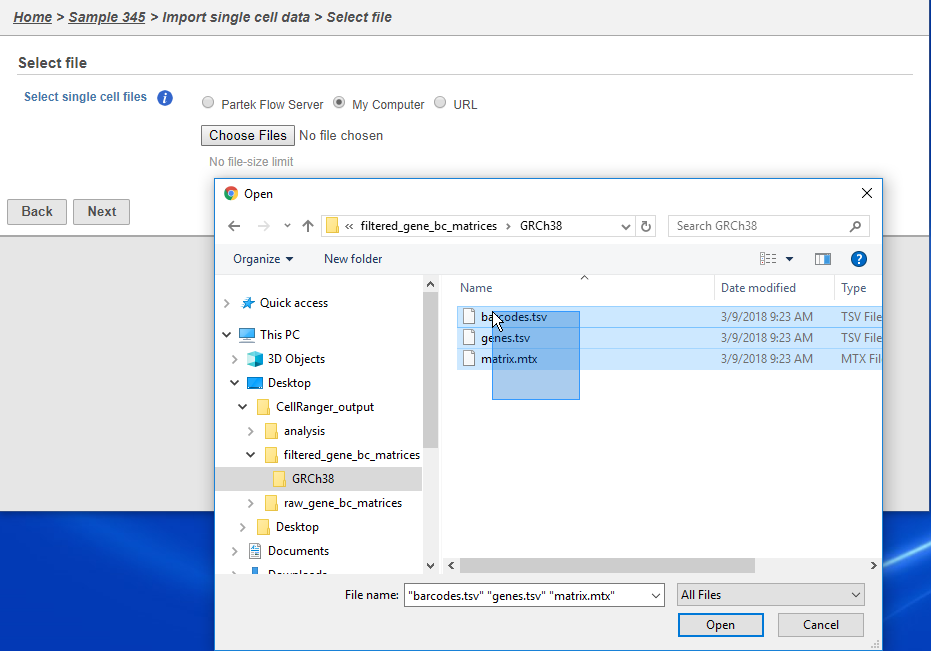
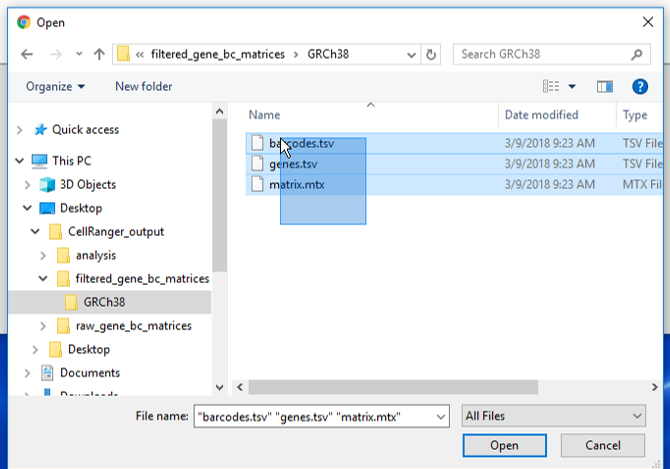
There are folders nested within the matrix folder, typically representing the reference genome it was aligned to. Navigate to the lowest subfolder, this should contain three files:
- barcodes.tsv
- genes.tsv
- matrix.mtx
Select all 3 files for importing import into Partek Flow
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
Specify the annotation file used when running the pipeline.
...