Page History
...
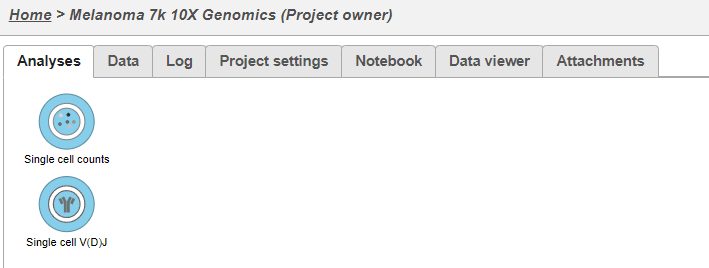
- This results in two starting nodes, one for single cell counts and one for single cell V(D)J as shown below. Note that once subsequent tasks are performed on a node, no more data can be imported into this project. The single cell counts node can be processed as usual; for help information related to this please see the tutorial for Analyzing Single Cell RNA-Seq Data.
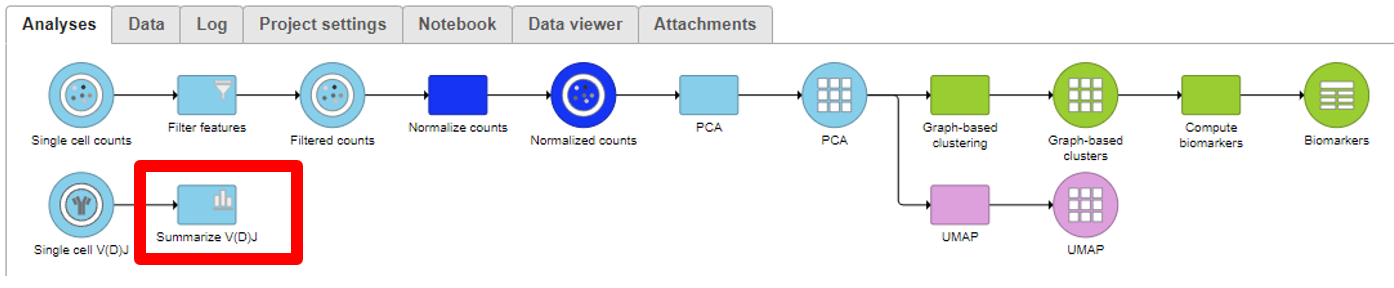
Analyzing the Single cell V(D)J node
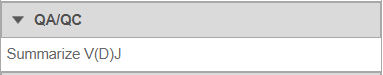
Summarize V(D)J
- Under QA/QC tasks is the Summarize V(D)J task which will summarize the V(D)J contents by Sample name, # Cells, Barcode count, Clonotypes, Variable genes, Diversity genes, Joining genes, and Constant genes.
- Double-click on the completed task to view the contents which can be downloaded.
Clonotype Frequency Plot
References
- Tonegawa, S. Somatic generation of antibody diversity. Nature 302,575–581 (1983). https://doi.org/10.1038/302575a0
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/output/annotation#contig-annotation
- https://support.10xgenomics.com/single-cell-vdj/software/overview/welcome
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/7.0/advanced/h5_matrices
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/7.0/output/matrices
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/using/vdj
- https://support.10xgenomics.com/single-cell-vdj/software/pipelines/latest/using/multi
Overview
Content Tools