Page History
...
To select a different layer, use the left mouse button to click on any node of the desired layer. All the nodes associated with the selected layer have the same color and when clicked will be displayed on the top of the stack.
Collapsing Tasks
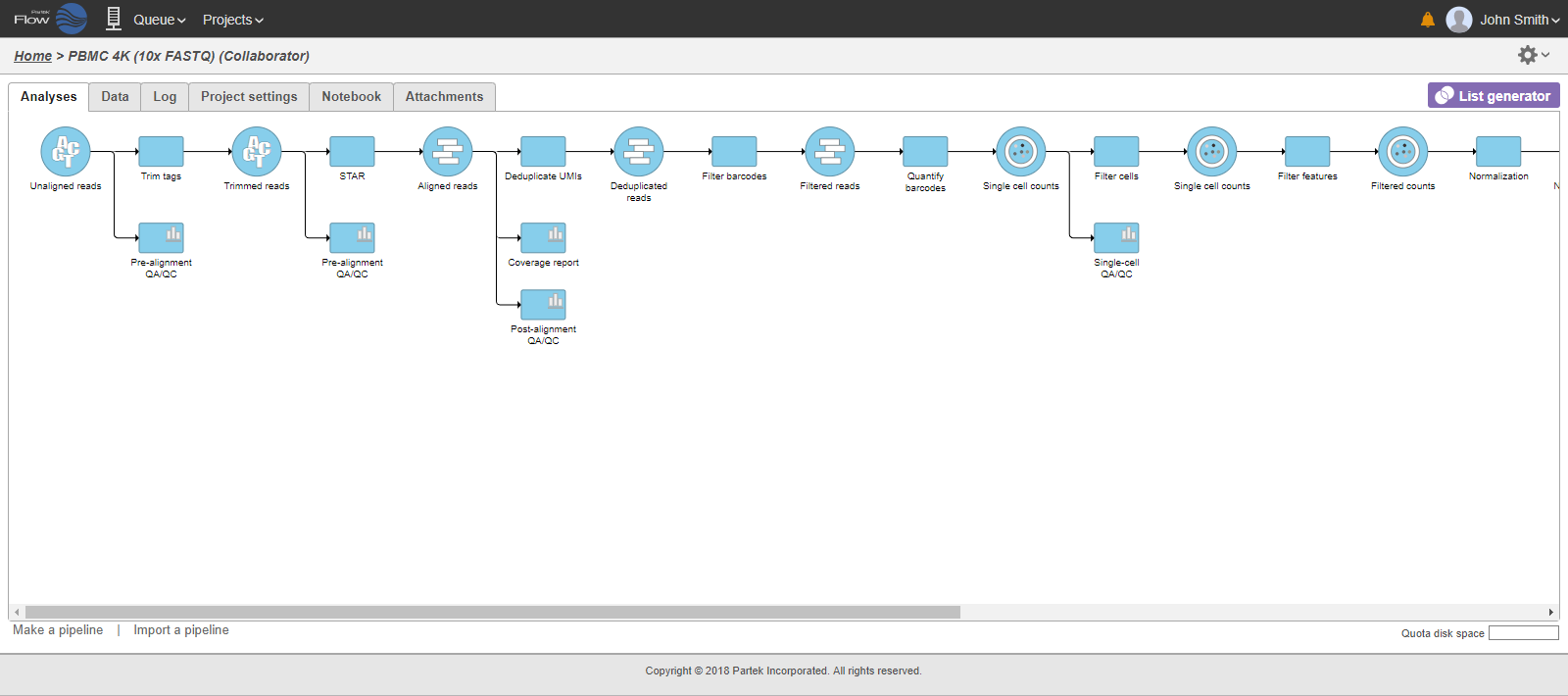
Addition of of task and respective resulting data nodes to project may lead to creation of long pipelines, extending well beyond the border of the canvas (Figure 18).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
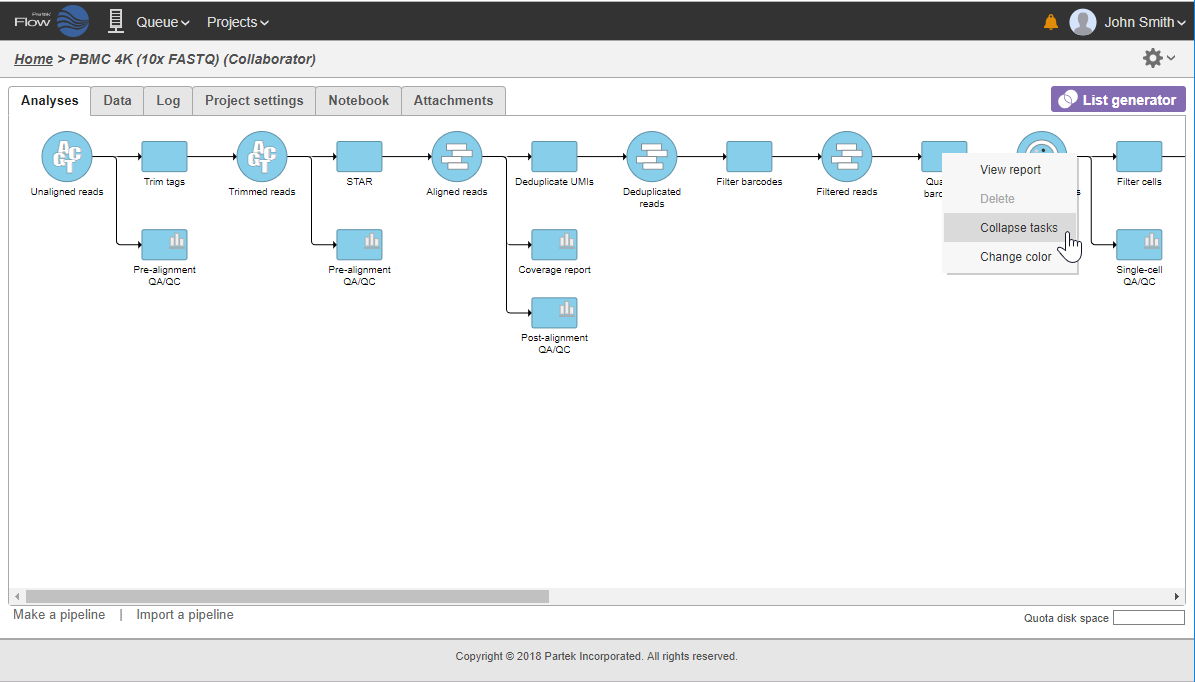
In that case, the pipeline can be collapsed, to hide the steps that are (no longer) relevant. For example, once the single-cell RNA-seq data has been quantified, Single cell counts data node will be a new analysis start point, as the subsequent analyses will not focus on alignment, UMI deduplication etc. To start, right-click on the task node which should become a boundary (=end) of the collapsed portion of your pipeline and select Collapse tasks (Figure 19).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
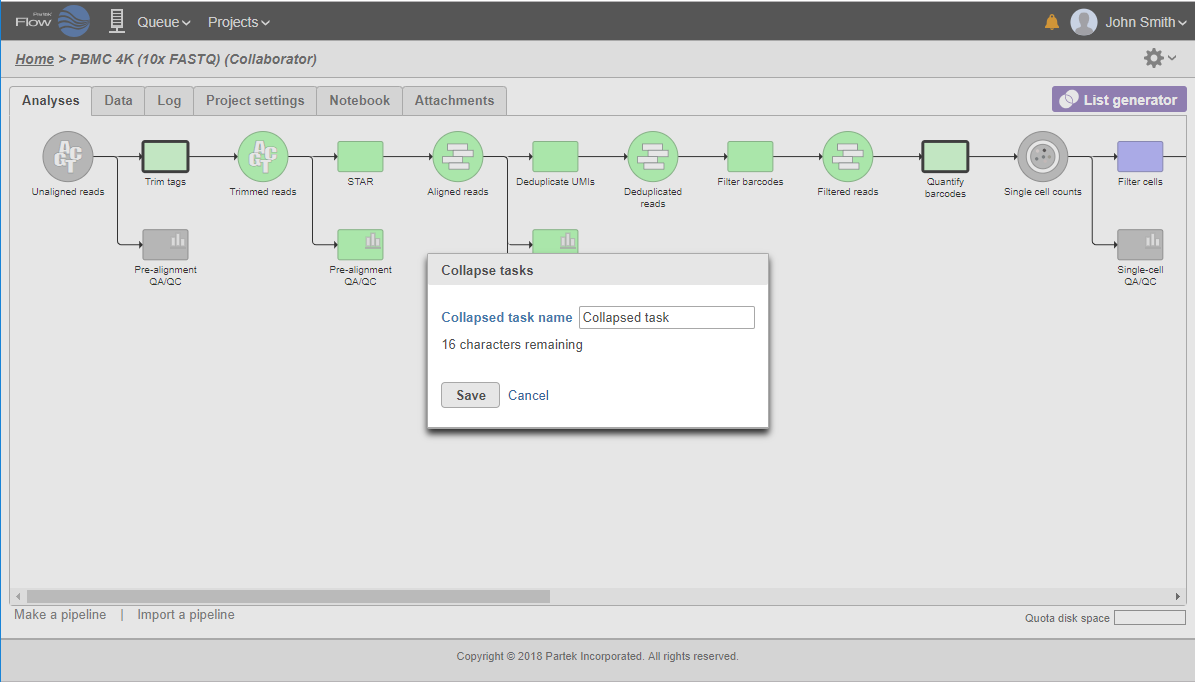
All the tasks on that layer will turn purple. Then left-click the task which should be the other boundary of the collapsed portion. The All the tasks that will be collapsed will turn green and a dialog will appear (Figure 19). In the example shown in Figure 19, the tasks between Trim tags and Quantify barcodes will be collapsed. Give the collapsed section a name (up to 30 characters) and select Save (Figure 20).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
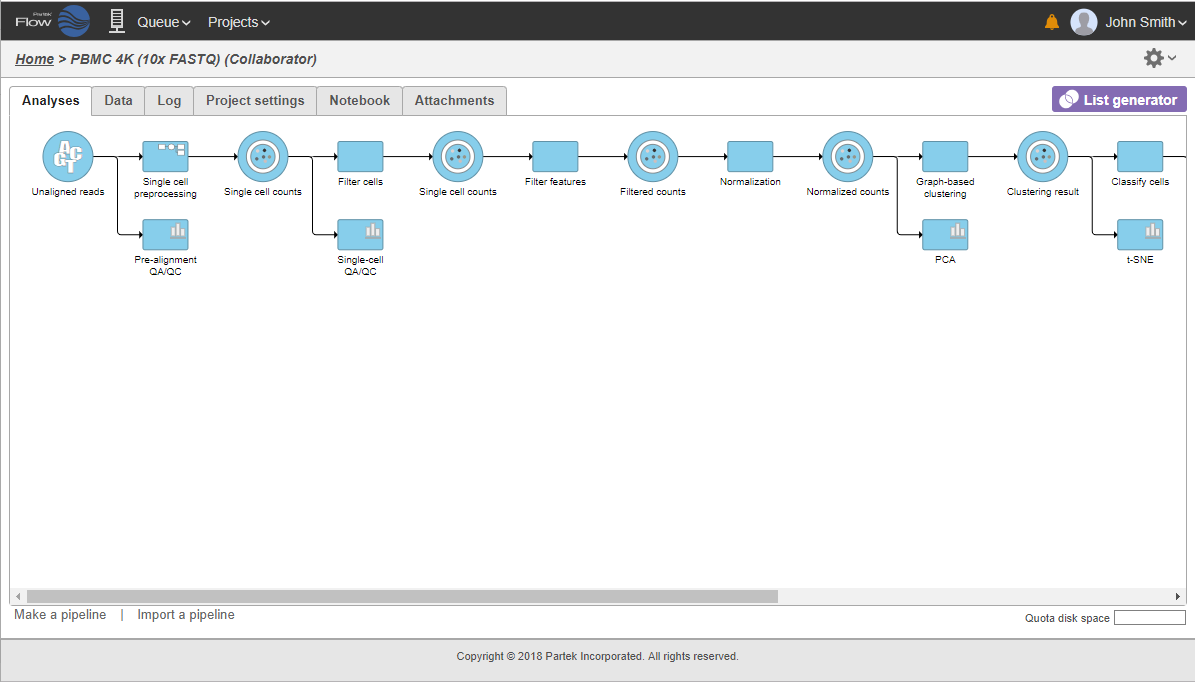
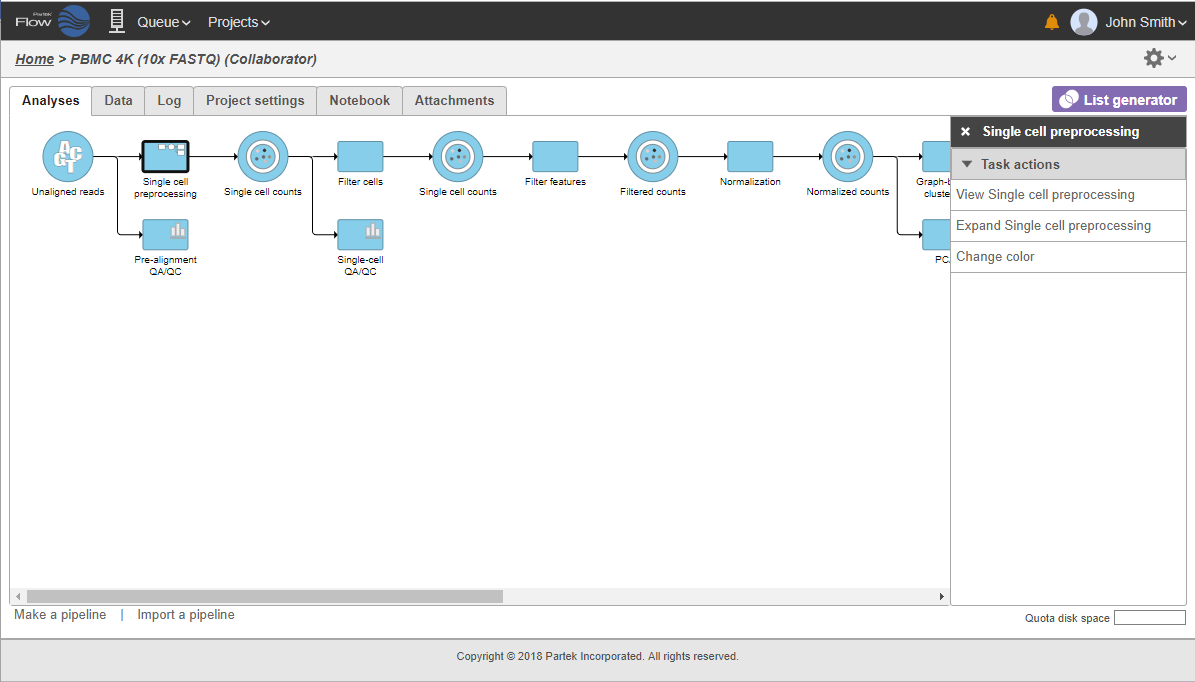
The collapsed portion of the pipeline is replaced by single task node, with a custom label ("Single cell preprocessing"; Figure 21). To re-expand the pipeline double click on the task node representing the collapsed portion of the pipeline.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To re-expand the pipeline double click on the task node representing the collapsed portion of the pipeline. Alternatively, single click on the node and select Expand... on the context-sensitive menu. Within the same menu, you can also preview the contents of the collapsed task by selecting View... (Figure 22).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Downloading Data
Data associated with any data node can be downloaded using the Download data link in the context sensitive menu (Figure 2223). Compressed files will be downloaded to the local computer where the user is accessing the Partek Flow server. Note that bigger files (such as unaligned reads) would take longer to download. For guidance, a file size estimate is provided for each data node. These zipped files can easily be imported by the Partek® Genomics Suite® software.
...
The lower right corner of the Analyses tab shows the Project disk space icon. Mouse over the icon to get information about the total space and usage available for your project (Figure 2324). The icon changes color from green to red as the used space gets closer to the total space (green < 80%, yellow < 90%, red >= 90%).
...