Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Pre-alignment QA/QC setup dialog is given in Figure 2. Examine reads allows you to control the number of reads processed by the tool; All reads, or a subset (One of every n reads). The latter option is obviously not as thorough, but is much faster than All reads.
...
The last control refers to .fastq files. Partek® Flow® can automatically detect the quality encoding scheme (Auto detect) or you can use one of the options available in the drop-down list. However, the auto-detection is only applicable for Phred+33 and Phred+64 type of quality encoding score. For early version of Solexa quality encoding score, select Solexa+64 from the Quality encoding drop down list. For a paired-end data, the pre-alignment QA/QC will be done on each read in pair separately and the results will be shown separately as well.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Most sequencing applications now use the phred quality score. This score indicates the probability that the base was accurately identified. The table below shows the corresponding base call accuracies for each score:
| Phred Quality Score | Base Call Accuracy |
|---|---|
| 10 | 90% |
| 20 | 99% |
| 30 | 99.9% |
| 40 | 99.99% |
The task report is organised in two tiers. The initial view shows project-level report with all the samples. An overview table is at the top, while matching plots are below.
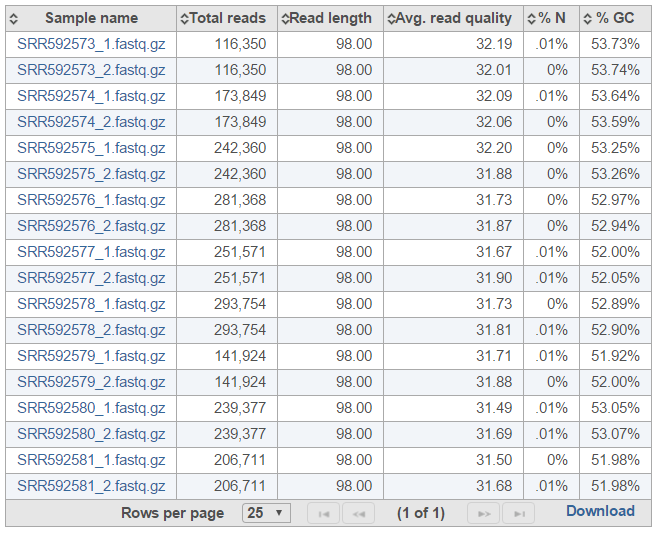
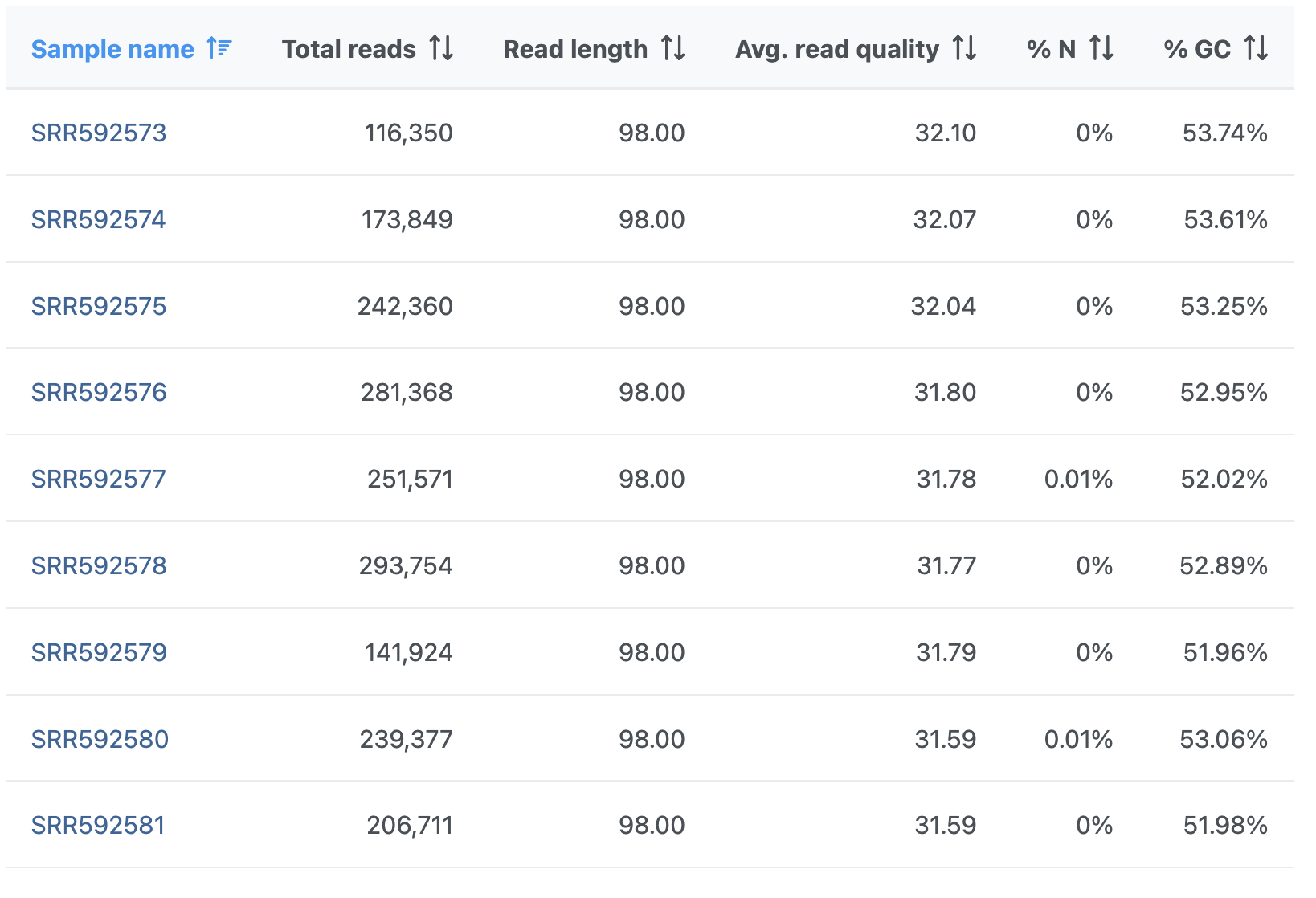
The Pre-alignment QA/QC output table contains one input file per row, with typical metrics on columns (%GC: fraction of GC content; %N: fraction of no-calls) (Figure 3). The file names are hyperlinks, leading to the sample-level reports. To save the table as a txt file to a local computer, push the Download link. Table columns can be sorted using double arrows icon ().
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
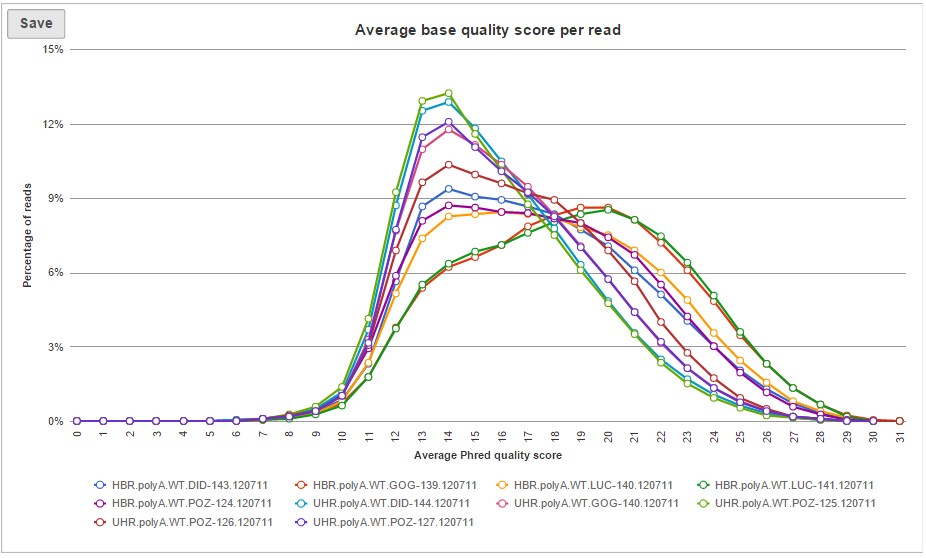
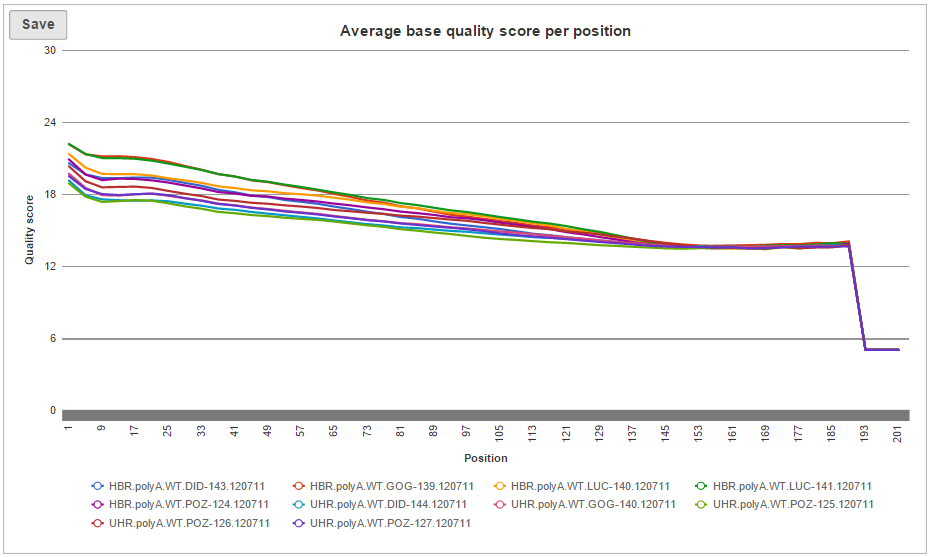
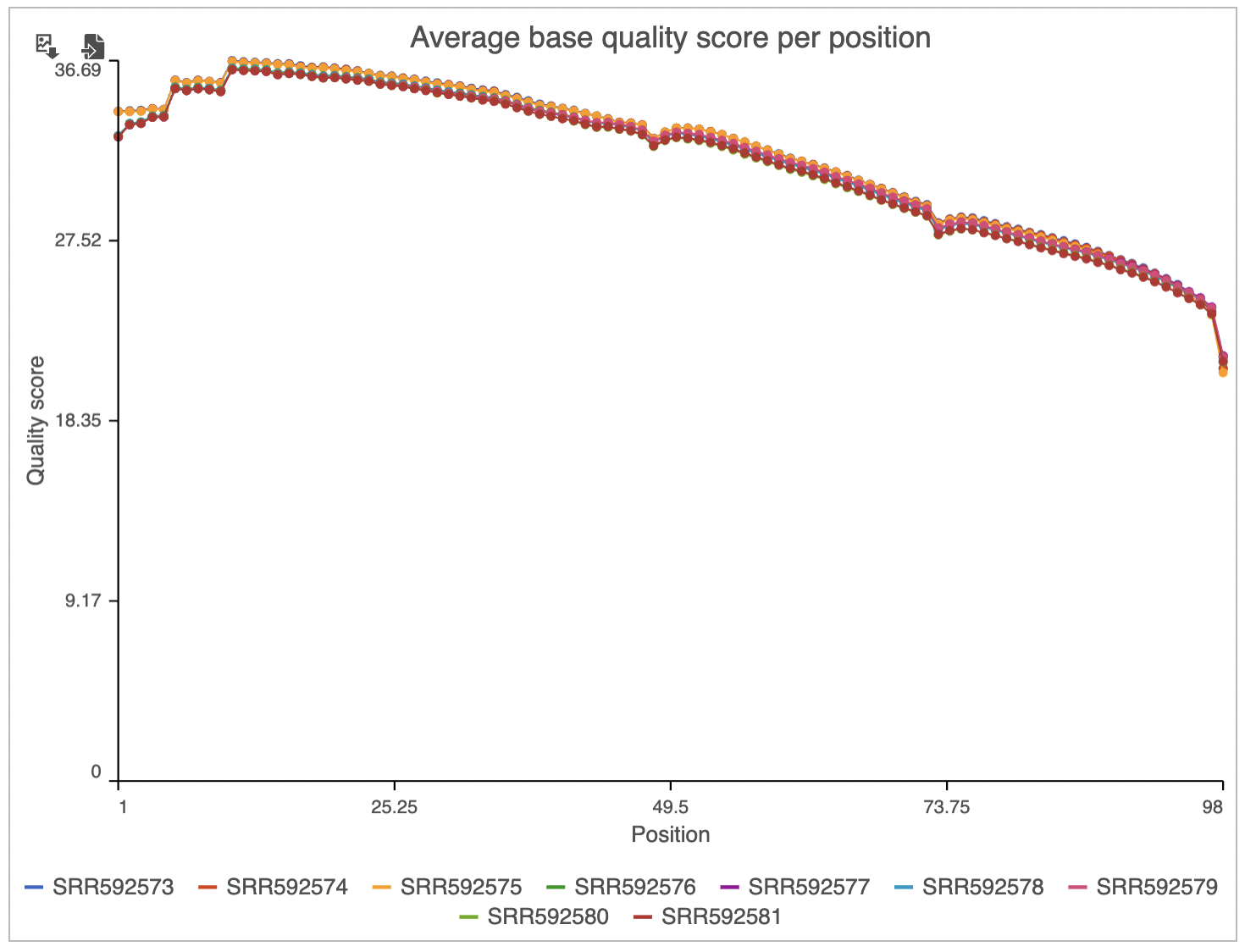
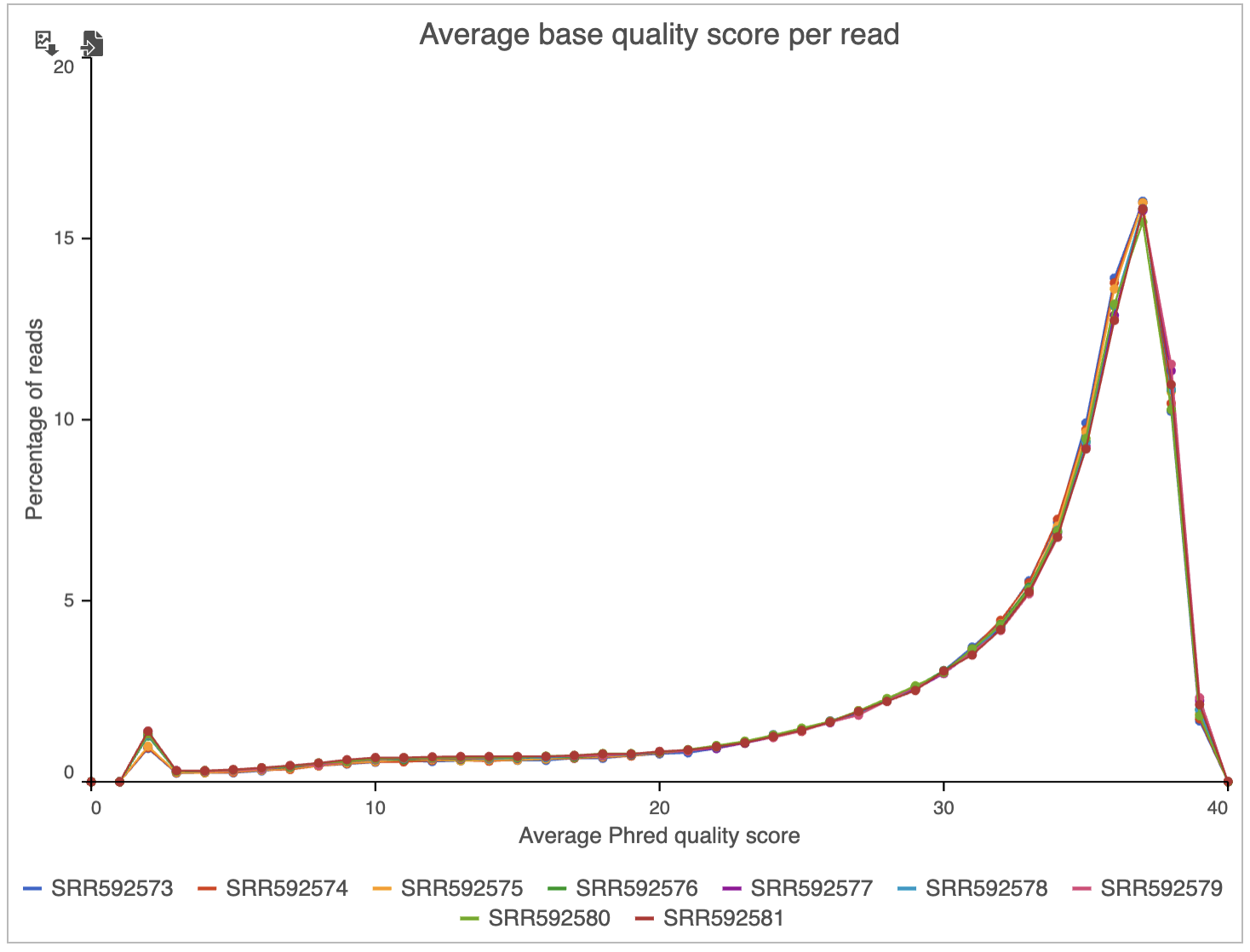
Two project-level plots are Average base quality per position and Average base quality score per read (Figure 4). The latter plot presents the proportion of reads (y-axis) with certain average quality score (meaning all the base qualities within a read are averaged; x-axis). Mouse over a data point to get the matching readouts. The Save icon saves the plot in a .svg format to the local machine. Each line on the plot represents a data file and you can select the sample names from the legend to hide/un-hide individual lines.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A sample-level report begins with a header, which is a collection of typical quality metrics (Figure 5).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Distribution of read lengths shows a single column for fixed length data (e.g. Illumina sequencing). However, for quality-trimmed data or non-fixed length data (like Ion Torrent sequencing), expect to see a read’s length distribution (Figure 7).
...
The pre-alignment QA/QC report as described above is generally available for the NGS data of fastq format. For other types of data, the report may differ depending on the availability of information. For example, for fasta format, there is no base quality score information and therefore all the figures or graphs related to base or read quality score will be unavailable.
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|