Page History
...
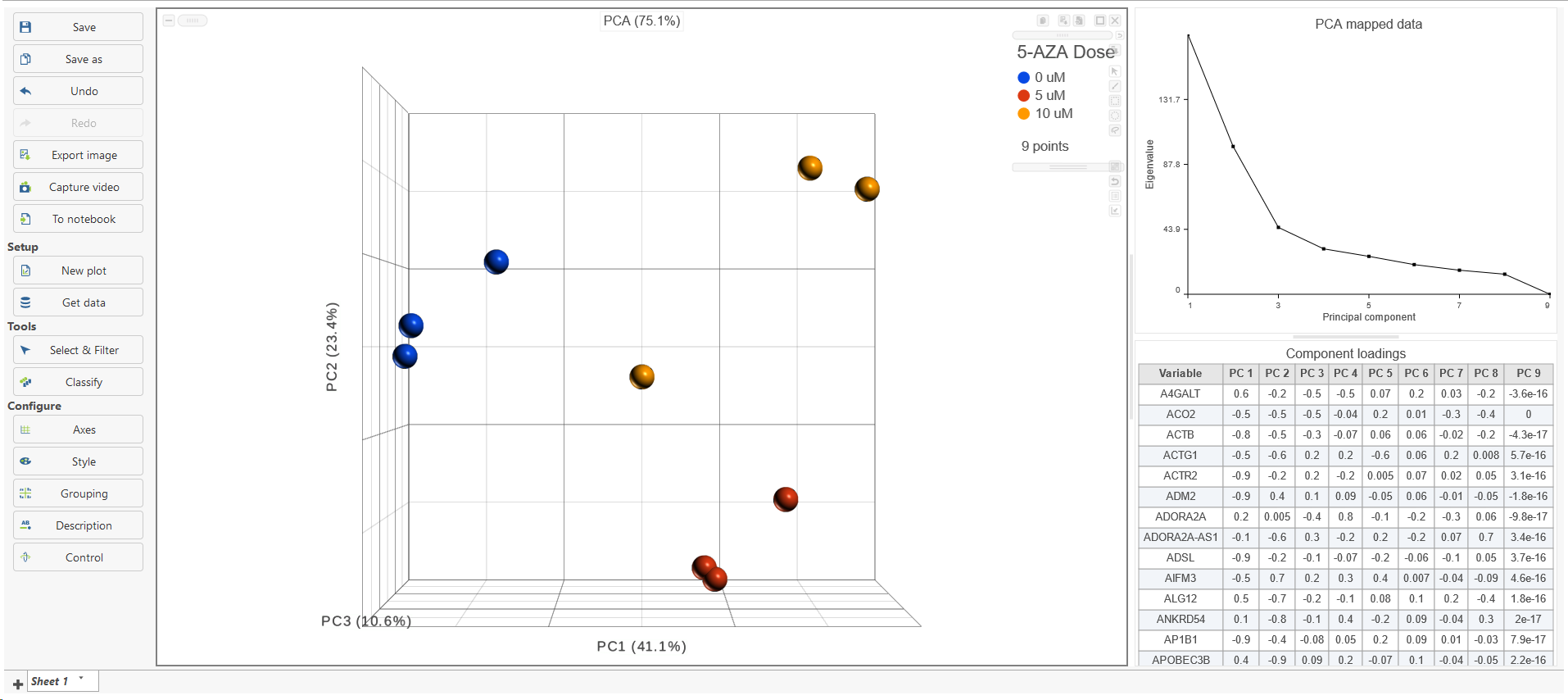
The PCA task creates a new task node, and to open it and see the result, do one of the following: select the PCA task node, proceed to the context sensitive menu and go to the Task result; or double-click on the PCA task node. The report containing eigenvalues, PC projections, component loadings, and mapping error information for the first three PCs.
When open the PCA node is opened in Data viewer, by default, it is the 3D scatterplot, Scree plot with Eigenvalues, and Component loadings table (Figure 2), each . Each dot on the 3D scatter plot represents an observation, while the first three PCs are shown on the X-, Y-, and Z-axis respectively, with the information content of an individual PC is in the parenthesis.
As an exploratory tool, the the PCA scatterplot is applied to view any groupings in the data set and generate hypotheses based on the outcome, or to spot possible outliers.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To rotate the 3D scatter plot left click & drag. To zoom in or out, use the mouse wheel. Click and drag the legend can move the legend to different location on the viewer.
Detailed configuration on PCA plot can be found by clicking Help>How-to videos>Data viewer section.
In the Data viewer, when a PCA data node is selected from Get Data under Setup (left panel), the node can be dragged and dropped to the screen (Figure 3), then you will have the option to select a scree plot and tables.
...