Page History
Introduction
Gene-specific analysis (GSA) is a statistical modeling approach used to test for differential expression of genes or transcripts in Partek Flow® Flow®. The terms "gene", "transcript" and "feature" are used interchangeably below. The goal of GSA is to identify a statistical model that is the best for a specific transcript, and then use the best model to test for differential expression.
Overview of GSA methodology
To test for differential expression, a typical approach is to pick a certain parametric model that can deliver p-values for the factors of interest. Each parametric model is defined by two components:
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
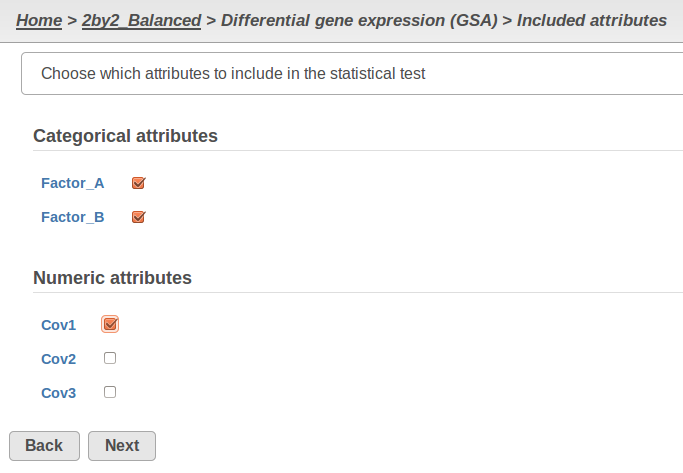
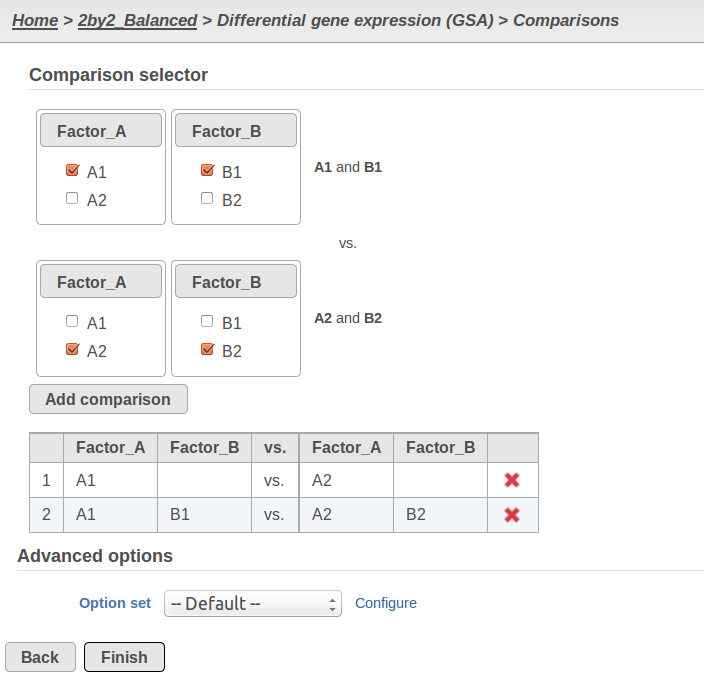
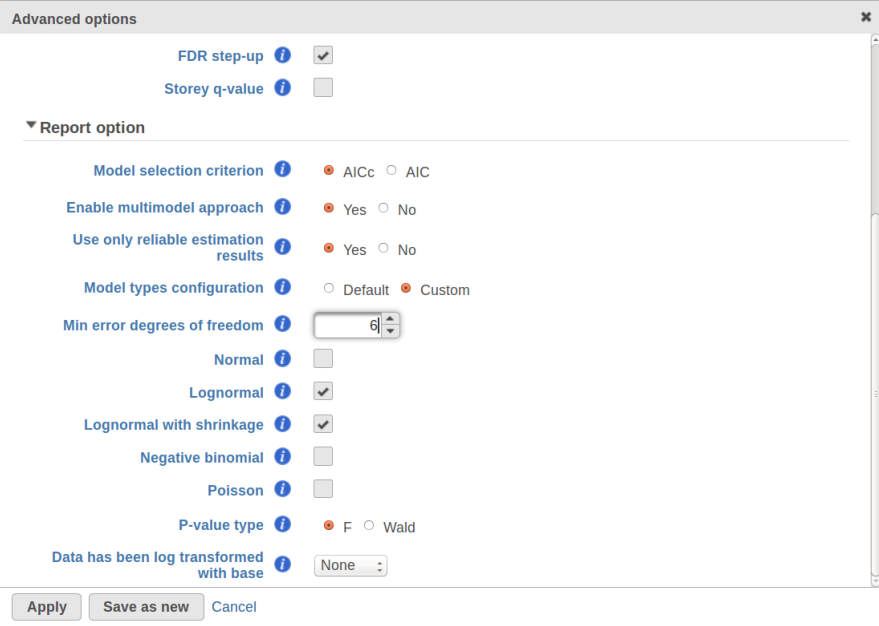
Currently, GSA is capable of considering the following five response distributions: Normal, Lognormal, Lognormal with shrinkage, Negative Binomial, Poisson (Figure 1). The GSA interface has an option to restrict this pool of distributions in any way, e.g. by specifying just one response distribution. The user also specifies the factors that may enter the model (Figure 2A) and comparisons for categorical factors (Figure 2B).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A) Choosing factors (attributes) in GSA B) Choosing comparisons in GSA |
...
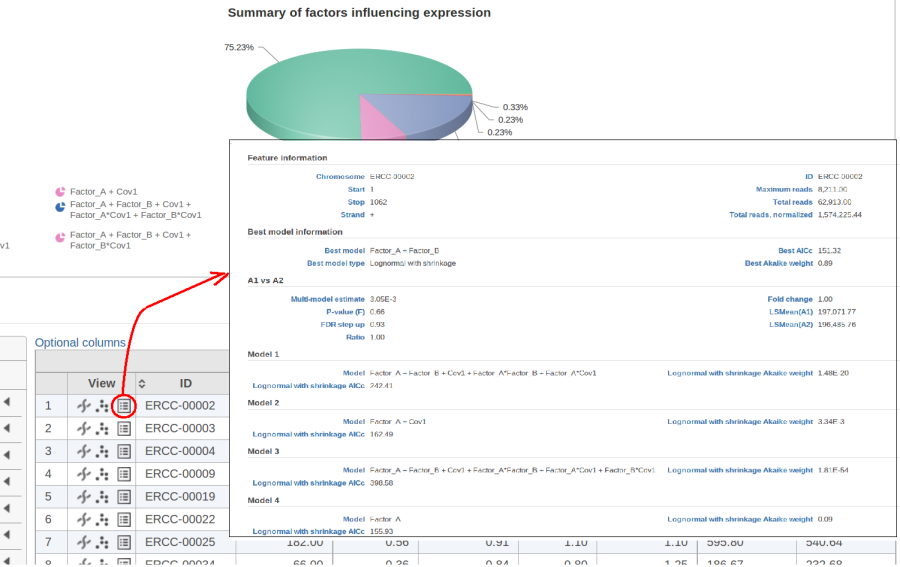
The model with the lowest information criterion is considered the best choice. It is possible to quantify the superiority of the best model by computing the so-called Akaike weight (Figure 3).The model's weight is interpreted as the probability that the model would be picked as the best if the study were reproduced. In the example above, we can obtain 15 Akaike weights that sum up to one. For instance, if the best model has Akaike weight of 0.95, then it is very superior to other candidates from the model pool. If, on the other hand, the best weight is 0.52, then the best model is likely to be replaced if the study were reproduced. We can still use this "best shot" model for downstream analysis, keeping in mind that that the accuracy of this "best shot" is fairly low.
| Numbered figure captions | ||||
|---|---|---|---|---|
|
...
Obtaining reproducible results in GSA
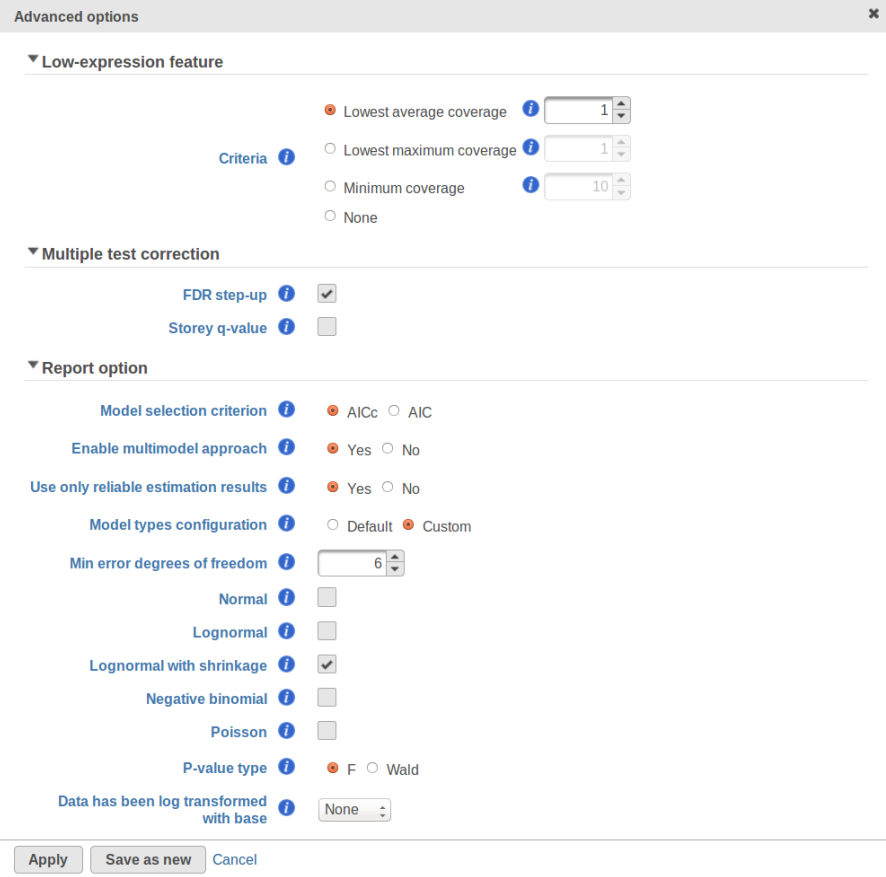
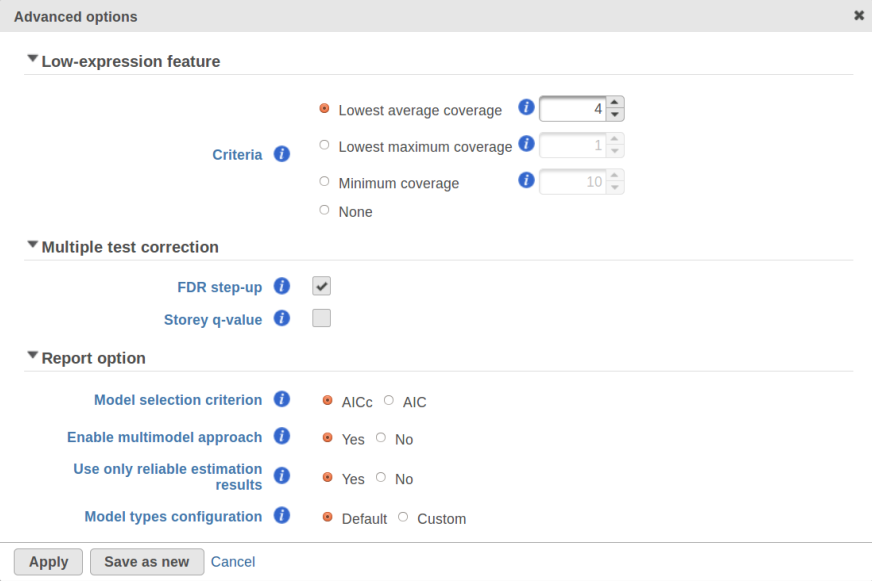
GSA is a great tool for automatic model selection: the user does not have to spend time and effort on picking the best model by hand. In addition, GSA allows one to run studies with no replication (one sample per condition) by specifying Poisson distribution in Advanced Options (Figure 1). However, not having enough replication is likely to deliver the results that are far more descriptive of the dataset at hand rather than the population from which the dataset was sampled. For instance, in the example from the previous section the number of fitted models (15) exceeds the number of observations (12), and therefore the results are unlikely to be representative of the population. If the study were to be reproduced with some 12 new samples, the list of significant features could be very different.
...
Second, the error degrees of freedom threshold in Default mode is adjusted automatically for the purpose of excluding designs that have too many terms compared to the number of observations. Third, the hierarchical design restriction explained in the previous section is also aimed at keeping the number of considered designs reasonable. The user can override the first two restrictions by switching from Default to Custom mode in Advanced Options (Figure 1). The 2nd order and hierarchical design restrictions cannot be disabled in GSA but they are absent in Flow ANOVA. The latter is similar to "Lognormal" option in GSA.
Using shrinkage plots
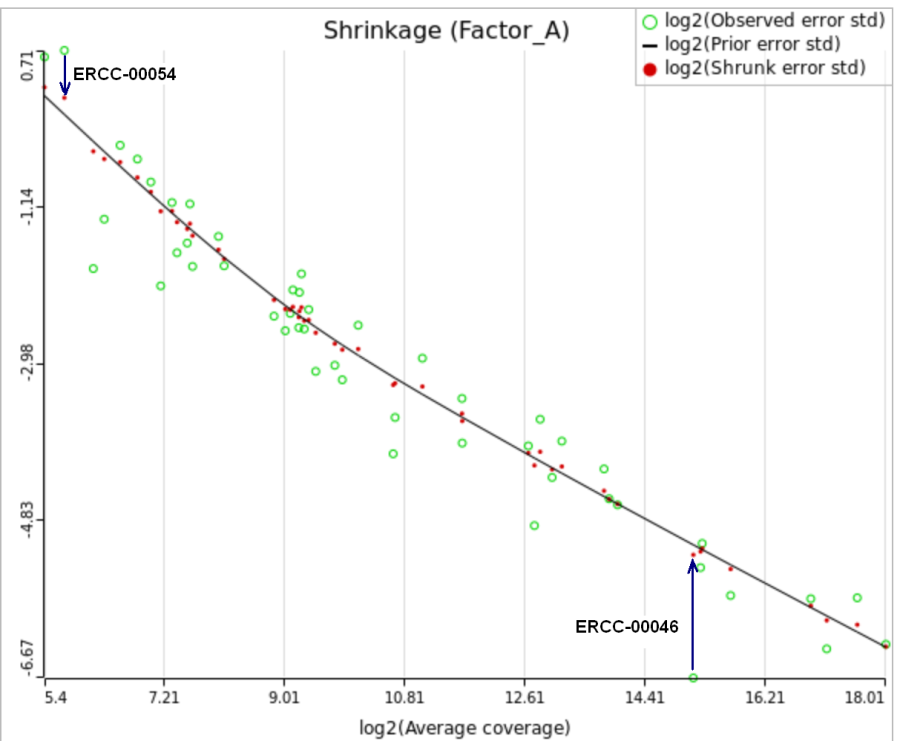
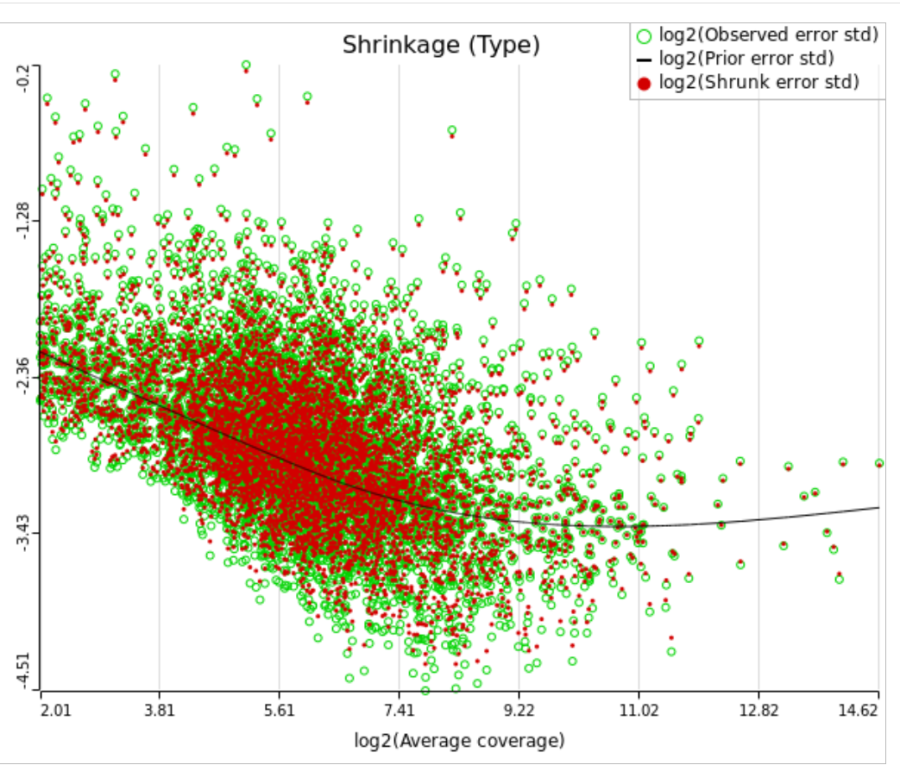
When "Lognormal with shrinkage" is enabled, a separate shrinkage plot is displayed for each design (Figure 4). First, a lognormal linear model is fitted for each gene separately, and the standard deviations of residual errors are obtained (green dots in the plot). Applying shrinkage amounts to two more steps. We look at how the errors change depending on the average gene expression and we estimate the corresponding trend (black curve). Finally, the original error terms are adjusted (shrunk) towards the trend (red dots). The adjusted error terms are plugged back into the lognormal model to obtain the reported results such as p-value.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
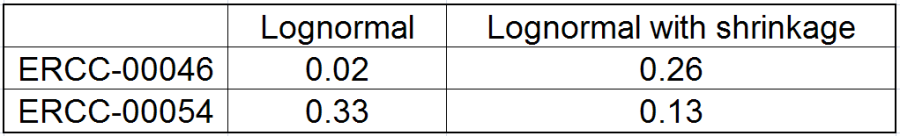
All other things being equal, the comparison p-value goes up as the magnitude of error term goes up, and vice-versa. As a result, the "shrunken" p-value goes up (down) if the error term is adjusted up (down). Table 1 reports some results for two features highlighted in Figure 4
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
For a large sample size, the amount of shrinkage is small, (Figure 5), and the "Lognormal" and "Lognormal with shrinkage" p-values become virtually identical.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
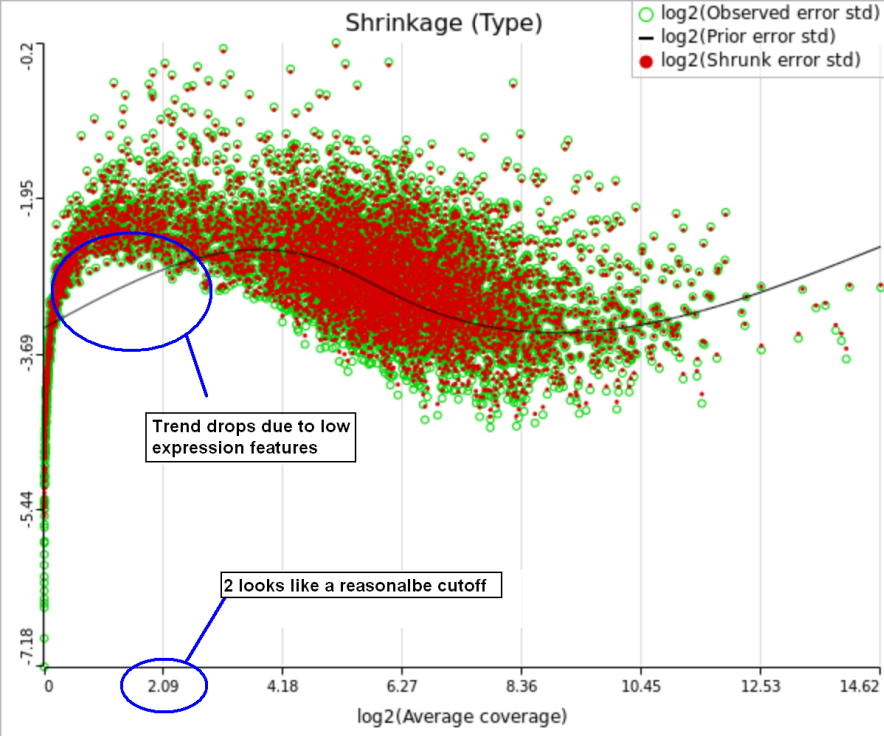
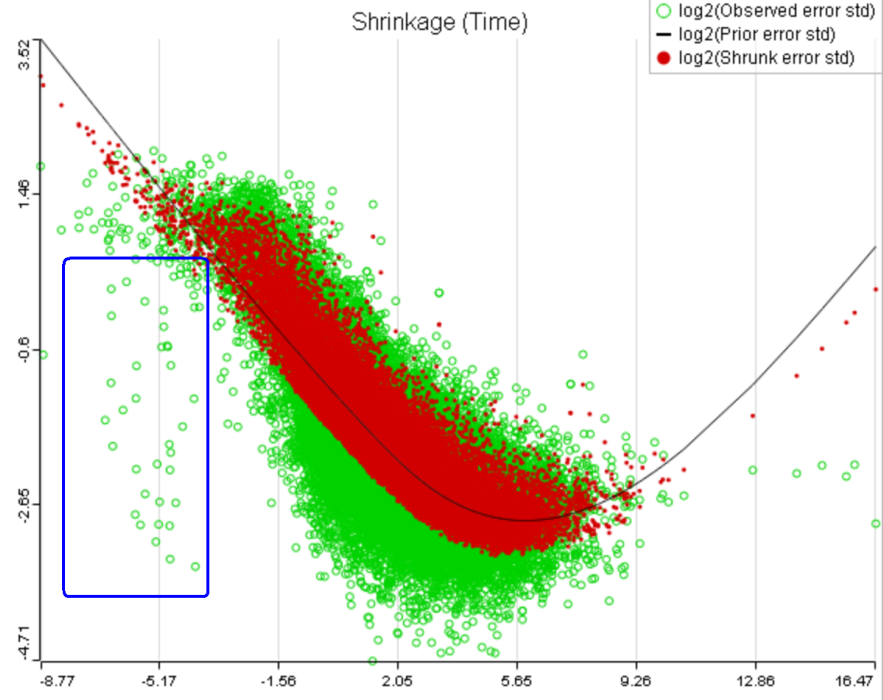
One important usage of the shrinkage plot is a meaningful setting of low expression threshold in Low expression filter section (Figure 6). For features with low expression, the proportion of zero counts is high. Such features are less likely to be of interest in the study, and, in any case, they cannot be modeled well by a continuous distribution, such as Lognormal. Note that adding a positive offset to get rid of zeros does not help because that does not affect the error term of a lognormal model much. A high proportion of zeros can ultimately result in a drop in the trend in the leftmost part of the shrinkage plot (Figure 56).
A rule of thumb suggested by limma authors is to set the low expression threshold to get rid of the drop and to obtain a monotone decreasing trend in the left-hand part of the plot.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
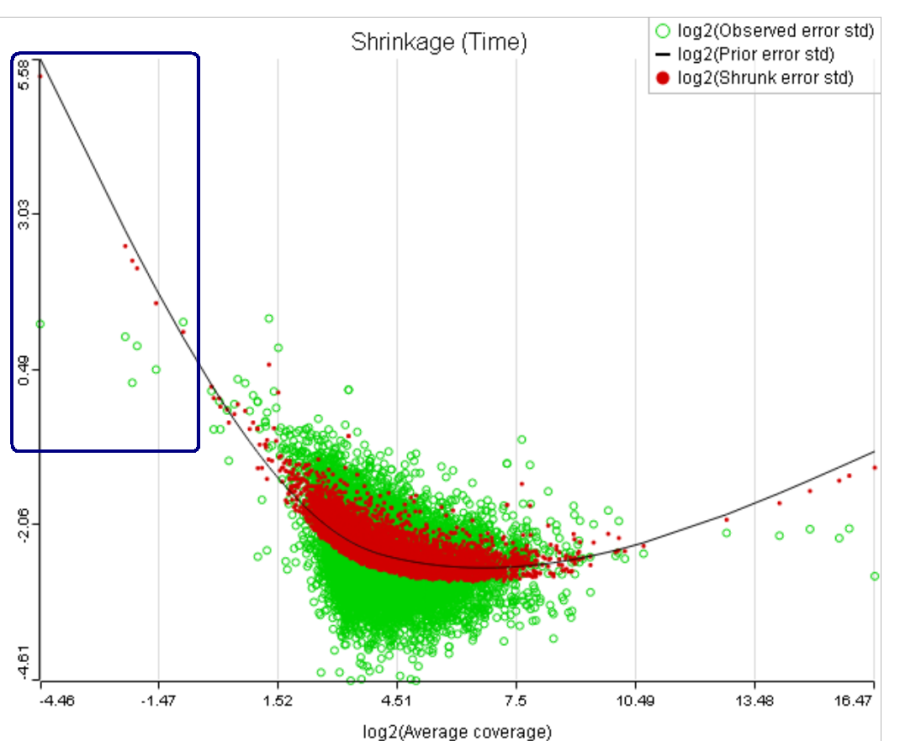
For instance, in Figure 5 it looks like a threshold of 2 can get us what we want. Since the x axis is on the log2 scale, the corresponding value for "Lowest average coverage" is 222^2=4 (Figure 6). After we set the filter that way and rerun GSA (Figure 7), the shrinkage plots takes the required form (Figure 78).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Note that it is possible to achieve a similar effect by increasing a threshold of "Lowest maximal coverage", "Minimum coverage", or any similar filtering option (Figure 67). However, using "Average coverage" is the most straightforward: the shrinkage procedure uses log2(Average coverage) as an independent variable to fit the trend, so the x axis in the shrinkage plot is always log2(Average coverage) regardless of the filtering option chosen in Figure 67.
As suggested above, Lognormal with shrinkage is not intended to work for a dataset that contains a lot of zeros, a set of low expression features being one example. In that case, a count-based (Poisson or Negative Binomial) model is more appropriate. With that in mind, it appears to be a simple solution to enable Poisson, Negative Binomial, and Lognormal with shrinkage in Advanced Options and let the model selection happen automatically. However, there are a couple of technical difficulties with that approach. First, shrinkage can fail altogether because of the presence of low expression features (in that case, the user will get an informative error message in GSA). Second, there is an issue of reproducibility discussed above. However, for a large sample size shrinkage becomes all but disabled and reproducibility is not a concern. In that case, one can indeed cover all levels of expression by using Poisson, Negative Binomial, and Lognormal together.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A) Average expression threshold can be raised to get rid of low expression features with abnormal error terms, circled in blue
B) Six low expression features (circled in blue) account for a very sharp increase in the trend which can have an unduly large effect on overall results |
...
Speaking of higher expression features, presently GSA has no automatic method to separate "abnormal" and "normal" features, so the user has to do some eyeballing of the shrinkage plot. However, for the purpose of investigating standalone outliers GSA can quantify the benefit of shrinkage in a well grounded way. In order to do that, one can enable both Lognormal and Lognormal with shrinkage in Advanced Options (Figure 9).
Figure 9:
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
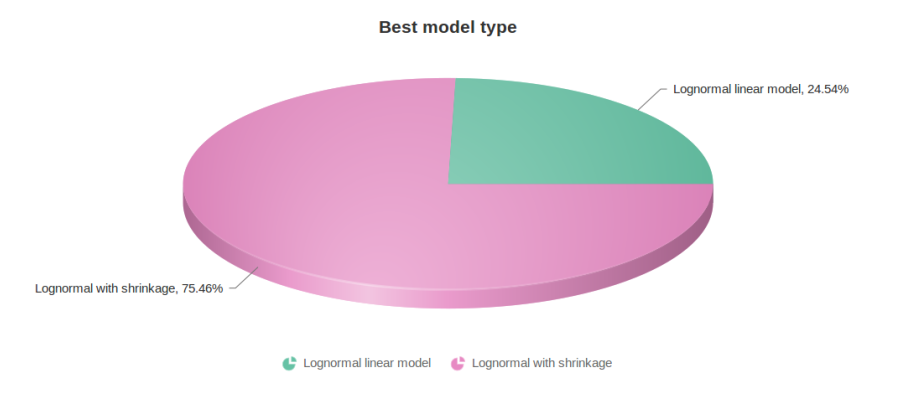
Figure 10 contains a pie chart for the dataset whose shrinkage plot is displayed in Figure 4. Because of a small sample size (two groups with four observations each) we see that, overall, shrinkage is beneficial: for an "average" feature, Akaike weight for feature-specific Lognormal is 25%, whereas Lognormal with shrinkage weighs 75%.
Figure 10:
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
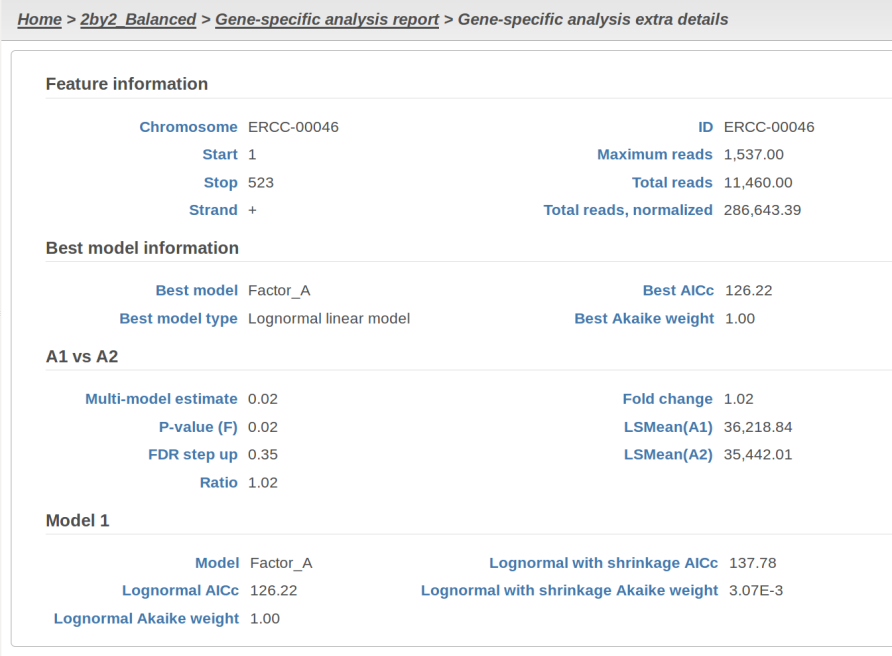
At the same time, if we look at ERCC-00046 specifically (Figure 11) we see that Lognormal with shrinkage fits so bad that its Akaike weight is virtually zero, despite having fewer parameters than feature-specific Lognormal.
Figure 11:
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Using multimodel inference appears to be a better alternative to the ad hoc method in DESeq2 that switches shrinkage on and off all the way. Once again, it is both technically possible and emotionally tempting to automate the handling of abnormal features by enabling both Lognormal models in GSA and applying them to all of the transcripts. Unfortunately, that can make the results less reproducible overall, even though it is likely to yield more accurate conclusions about the drastically outlying features.
Additional Assistance
If you need additional assistance, please visit partek.com/PartekSupport to submit a help ticket or find regional phone numbers to call Partek support.
References
[1] Auer, 2011, A two-stage Poisson model for testing RNA-Seq.
[2] Burnham, Anderson, 2010, Model selection and multimodel inference.
[3] Dillies et al, 2012, A comprehensive evaluation of normalization methods.
[4] Storey, 2002, A direct approach to false discovery rates.
[5] Law et al, 2014, Voom: precision weights unlock linear model analysis. Note that this paper introduces both "limma trend" and "limma voom", but the present implementation in GSA corresponds to "limma trend".
[6] Love et al, 2014, Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2.
[7] Bioconductor support forum. Accessed last: 4/12/16. {+} https://support.bioconductor.org/p/80745/#80758+
| additional- |
|---|
Copyright © 2016 by Partek Incorporated. All Rights Reserved. Reproduction of this material without express written consent from Partek Incorporated is strictly prohibited.
| assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|