Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
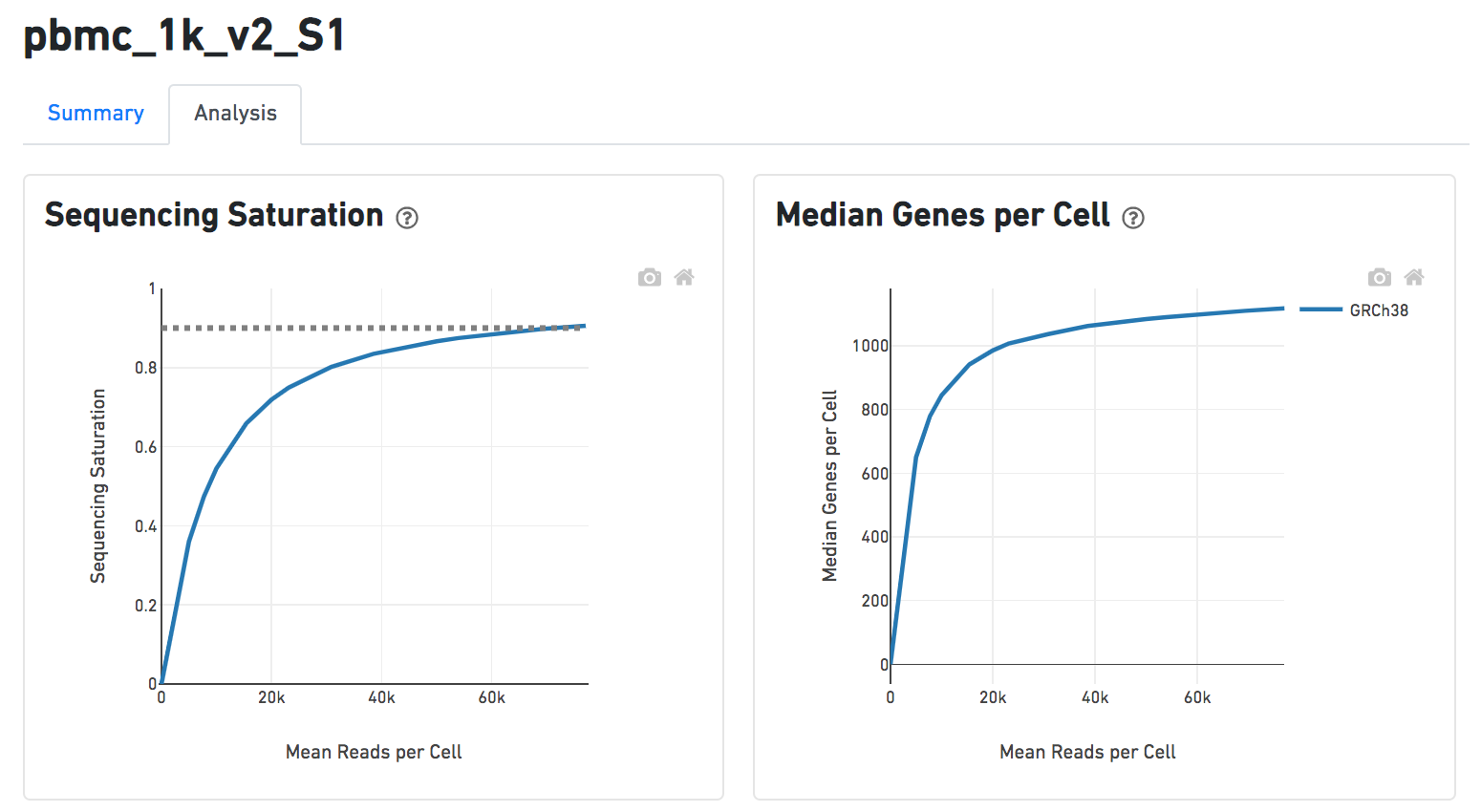
Another two plots -biplots of Sequencing Saturation and Median Genes per Cell to Mean Reads per Cells have been included on the Analysis report as they are important metrics to the library complexity and sequencing depth (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
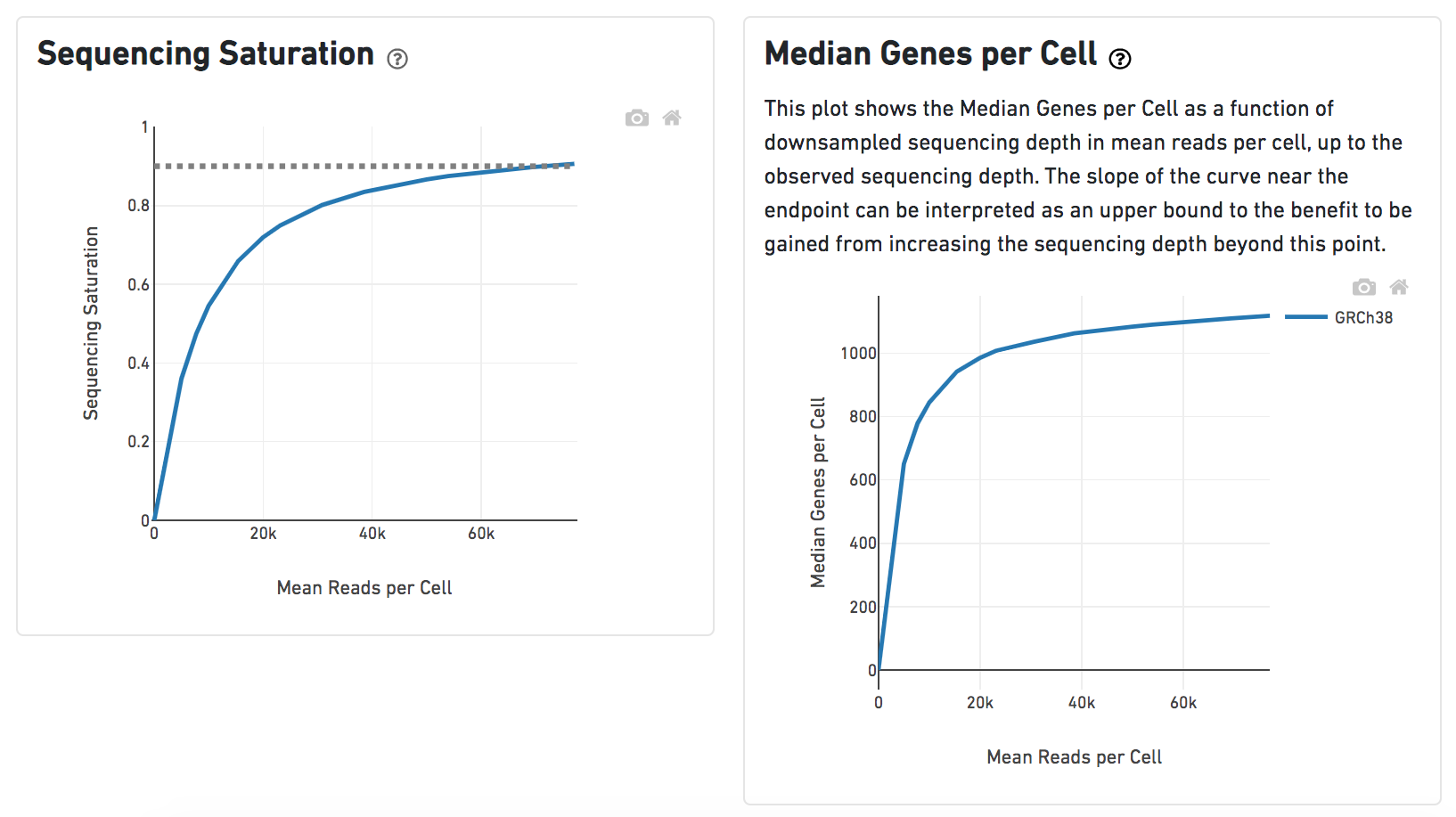
A lot of details would be exhibited and the panel would be expanded correspondingly if the the symbolin the panel on each report has been clicked. In the example below, the plot of Median Genes per Cell has been expanded while the Sequencing Saturation plot hasn't (Figure 9).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
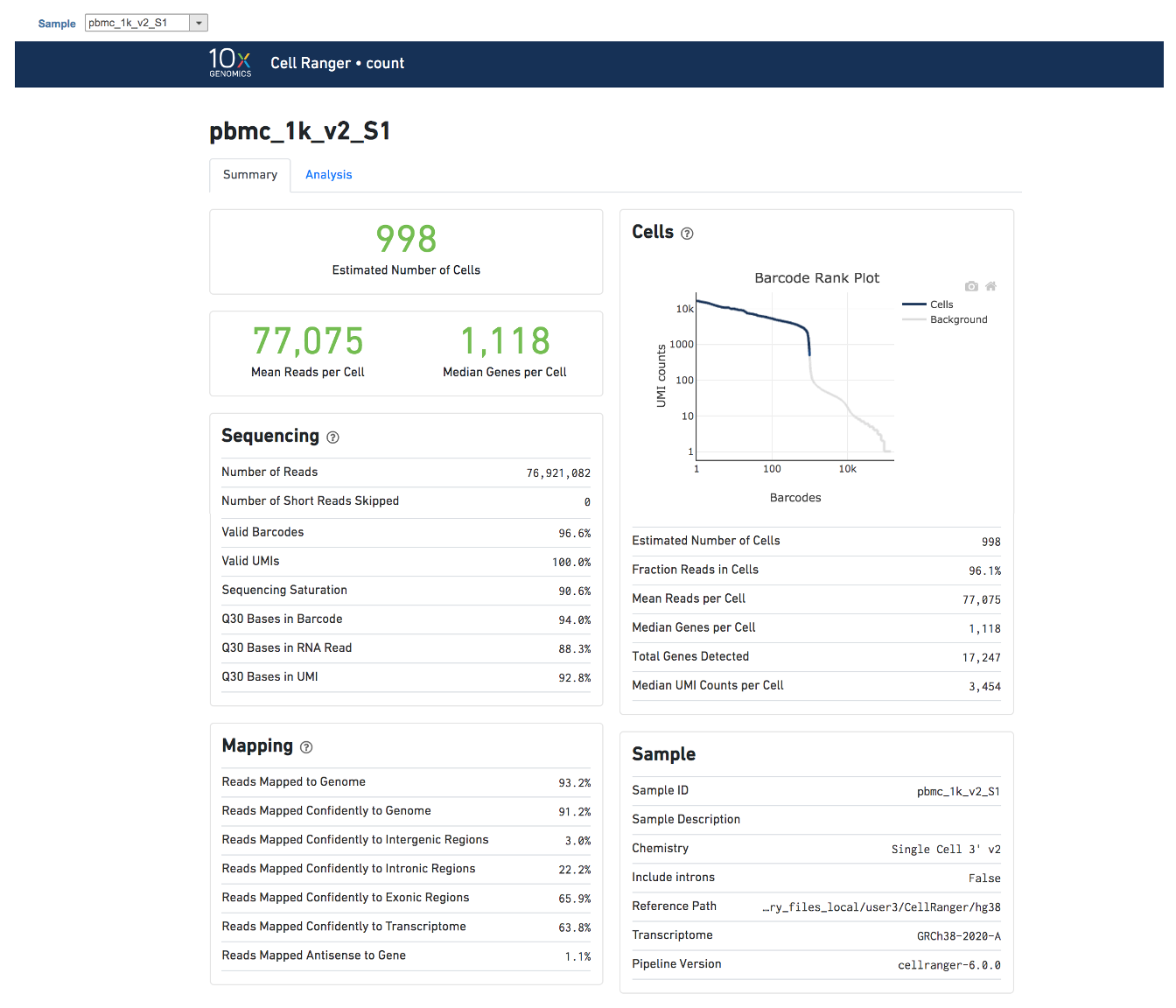
Users can click Configure to change the default settings In Advanced options (Figure 5).
Expected cells: Expected number of recovered cells. Default: 3,000 cells.
Force cells: Force pipeline to use this number of cells, bypassing the cell detection algorithm. Use this if the number of cells estimated by Cell Ranger is not consistent with the barcode rank plot.
References
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/6.0/release-notes
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/4.0/release-notes
...