Page History
...
The report produced by DESeq2 is similar to the ANOVA report; each row is a feature and columns include p-value, FDR p-value, and fold change in linear scale for each contrast. However DESeq2(R) report doesn't have LSMeans information on the compared groups.
Handling of special cases in DESeq2
The R implementation of DESeq2 and DESeq2(R) fail to generate results in some special cases of input data that are handled differently in Partek's implementation of DESeq2.
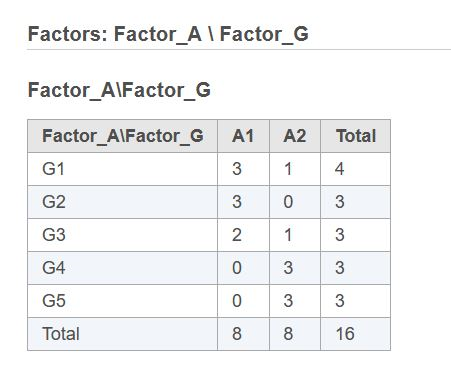
First, if two or more categorical factors are present in the model, there can be a situation when some combinations of factor levels have no observations. In Flow, one can see if that is the case by clicking "Cross tabulation" link after the factors have been selected:
In this example, if the user tries to fit the model including Factor_A, Factor_G, and Factor_A*Factor_G terms, R implementation of DESeq2 fails. At the same time, none of these three terms is completely redundant, even though not all contrasts are estimable. It is possible to obtain an answer in R only by combining Factor_A and Factor_G into a single new factor with no empty levels. Partek's implementation of DESeq2 eliminates the need for that extra step and produces an answer right away. However, the results (most likely, p-values) may be somewhat different from what one would obtain by using a combined factor in R. To match R results perfectly, one has to create a combined factor in Flow also.
Second, occasionally all of the gene-wise dispersion estimates are too close to zero. In that case, R implementation fails with an error message "all gene-wise dispersion estimates are within 2 orders of magnitude from the minimum value ... one can instead use the gene-wise estimates as final estimates". As suggested in the error message, Flow implementation uses gene-wise dispersion estimates instead of failing the task.
In all of the special cases, an informative warning is output in Flow log.
References
Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014;15(12): 550.
...