Page History
...
Hierarchical clustering is an unsupervised technique, meaning that the number of clusters is not specified up frontupfront. In the beginning, each row and/or column is considered a cluster. The two most similar clusters are combined and continue to combine until all objects are in the same cluster. Hierarchical clustering produces a tree (called a dendrogram) that shows the hierarchy of the clusters.
...
| Table of Contents | ||
|---|---|---|
|
Invoking Hierarchical Clustering
To invoke hierarchical clustering, select a Quantification data node (to cluster samples) containing count data (e.g. Gene counts, Normalized counts, Single cell counts), or a Feature list data node (to cluster significant genes/transcripts) and then click on the Hierarchical clustering / heat map option in the context sensitive menu (Figure 1).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
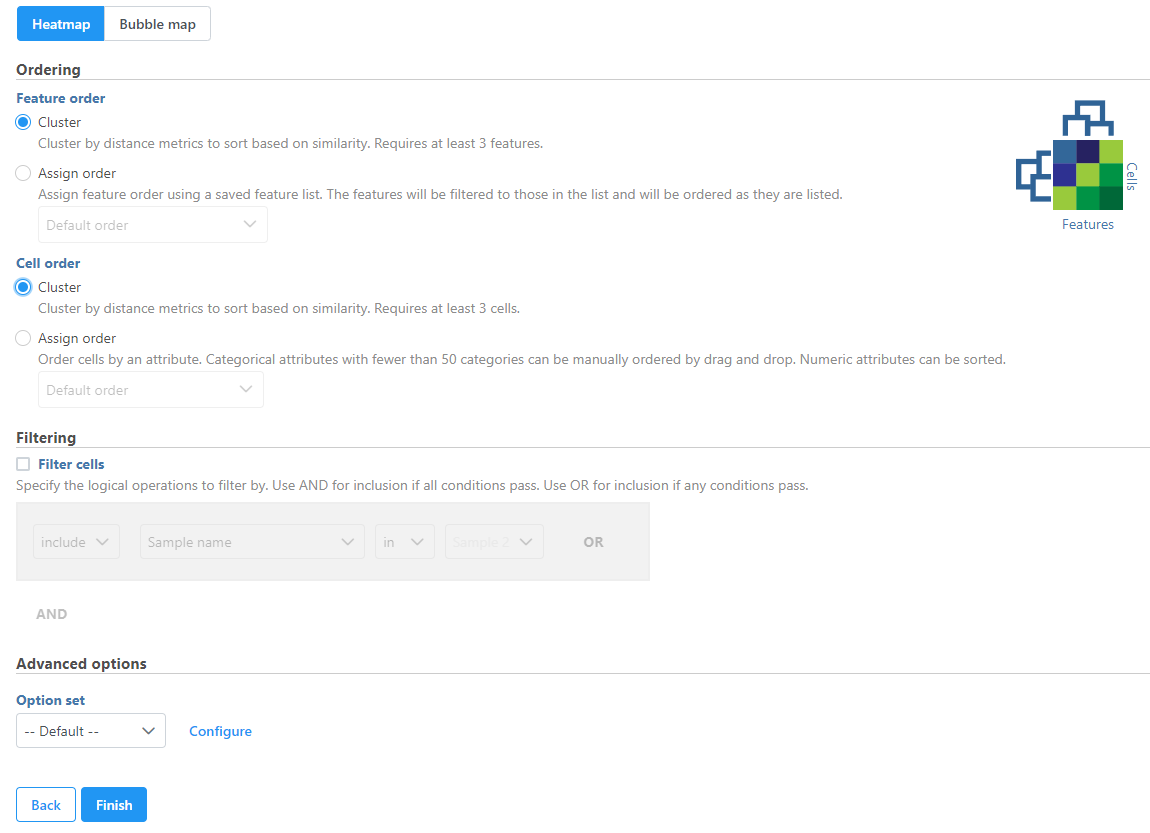
The hierarchical clustering setup dialog (Figure 2) enables you to control the clustering algorithm. Starting from the top, you can choose to Cluster samples, Cluster features plot a Heatmap or a Bubble map (clustering can be performed on both plot types). Next, perform Ordering by selecting Cluster for either feature order (genes/transcripts/proteins) or cell/sample/group order or both. Note the context-sensitive image that helps you decide to either perform hierarchical clustering (dendrogram) or assign order (arrow) for the columns and rows to help you orient yourself and make decisions (In Figure 2 below, Cluster is selected for both . By default, if there are less than 3000 samples, the Cluster samples check button is selected, if there are less than 3000 features, the Cluster features check button is selected. Otherwise the check button is de-selected.options so a dendrogram is shown in the image).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
When choose Assign order, the Default order of cells/samples/groups (rows) is based upon the labels as displayed in the Data tab and features (columns) are dependent on the input data of the data node.
Feature order can be assigned by selecting a managed list (e.g. generate saved feature lists from report nodes or add lists under list management in the settings) in the drop-down which will limit the features to only those in the list and the features will be ordered as they are listed. If a feature is not available, based on the input of the data node, it will not be shown in the plot (in other words, if the features from the list are not there they will not be plotted). Note that If no features are available from the data node, the task will not be able to perform and an error message will be shown.
Cell/Sample/Group order can also be assigned by choosing an attribute from the drop down list. Click and drag to rearrange categorical attributes; numeric attributes can be sorted in ascending or descending order (note the arrows in the image which are different from the dendrogram for Cluster) (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Another way to invoke a heatmap without performing clustering is via the data viewer. When you select the Heatmap icon in the available plots list, data nodes that contain two-dimensional matrices can be used to draw this type of plot. A bubble map can also be similarly plotted (use the arrow from the heatmap icon to select a Bubble map for descriptive statistics that have been generated in the data analysis pipeline.
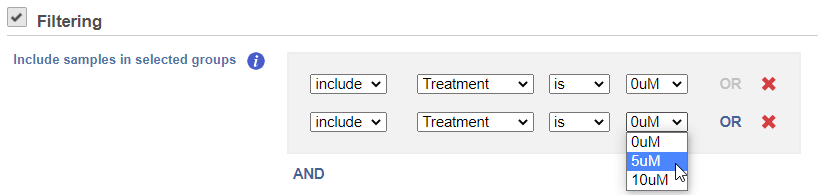
If you do not want to cluster all the samples, but select a subset based on a specific sample or cell attribute (i.e. group membership), check Filter cells under Filtering and set a filtering rule using the drop down lists (Figure 4). Notice the drop-down lists allow more than one factor (when available) to be selected at a time. When configuring the filtering rule, use AND to ensure all conditions pass for inclusion and use OR for any conditions to pass.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| |||
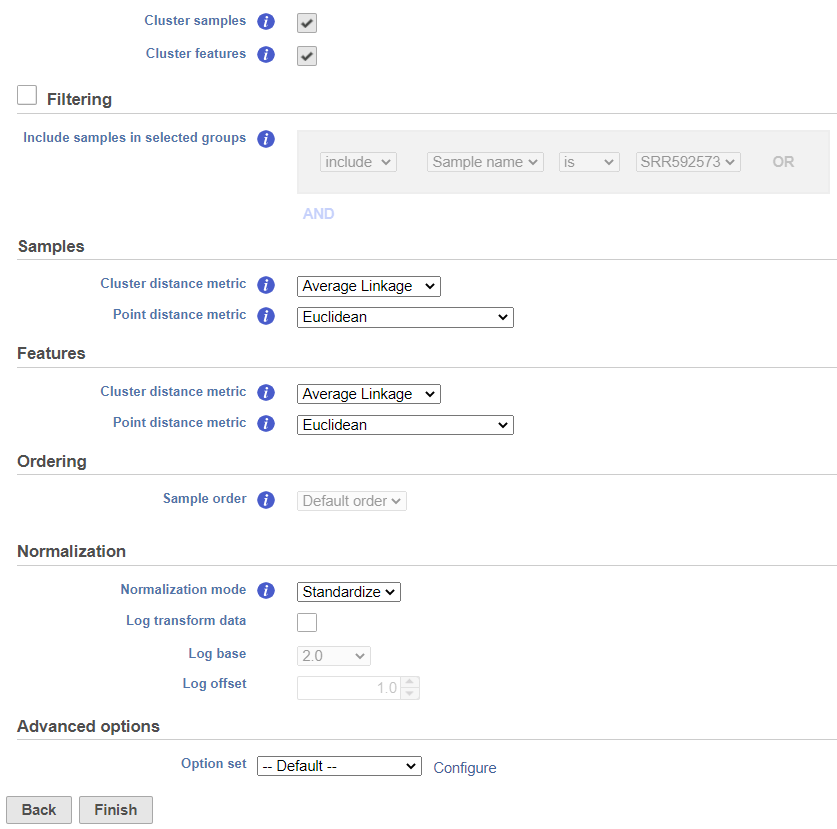
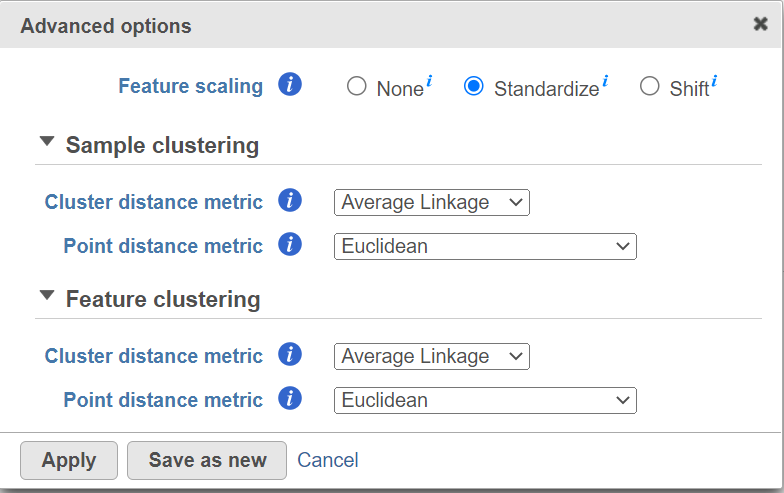
Hierarchical clustering uses distance metrics to sort based on similarity and is set to Average Linkage by default. This can be adjusted by clicking Configure under Advanced options (Figure 5). You can choose how the data is scaled (sometimes referred to as normalized). There are three Feature scaling options, Standardize (default for a heatmap) will make each column mean as zero and standard deviation as 1 in all features. This is the default scaling for a heatmap and it makes all of the features (e.g., genes or proteins) have equal weight; standardized values are also known as Z-scores. The scaling mode Shift will make each column mean as zero. Choose None to not scale and perform clustering on the values in the input data node (this is the default for a bubble map). If a bubble map is scaled, scaling will be performed on the group summary method (color).
Cluster distance metric for cells/samples and features is used to determine how the distance between two clusters will be calculated. :
- Single Linkage: the distance between two clusters is determined by the distance of the closest objects in the two clusters
...
- Complete Linkage: the distance between two clusters is equal to the distance between the two furthest members of those clusters
...
- Average Linkage: the average distance between all the pairs of objects in the two different clusters is used as the measure of distance between the two clusters
...
- Centroid method: the distance between two clusters is equal to the distance between the centroids of those clusters
...
- Ward's method: the distance between two clusters is designed to minimize the size of an error measure based on the sum of squares
...
Point distance metric is used to determine the distance between two rows or columns. For a thorough discussionmore detailed information about the equations, we refer you to the distance metrics chapter.
If you do not want to cluster all the samples, but select a subset based on specific sample attributes (i.e. group membership), use the Filtering drop down list (Figure 3). The default value of the Filtering option is All samples.
You can choose to how the data is normalized. Under the Normalization mode dropdown, Standardize (default) will make each column mean as zero and standard deviation as 1 in all features. This is the default normalization and it makes makes all the features (e.g., genes) have equal weight. The normalization mode Shift will make each column mean as zero. Choose None to perform clustering on the values in the quantified data node.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| |||
Heatmap
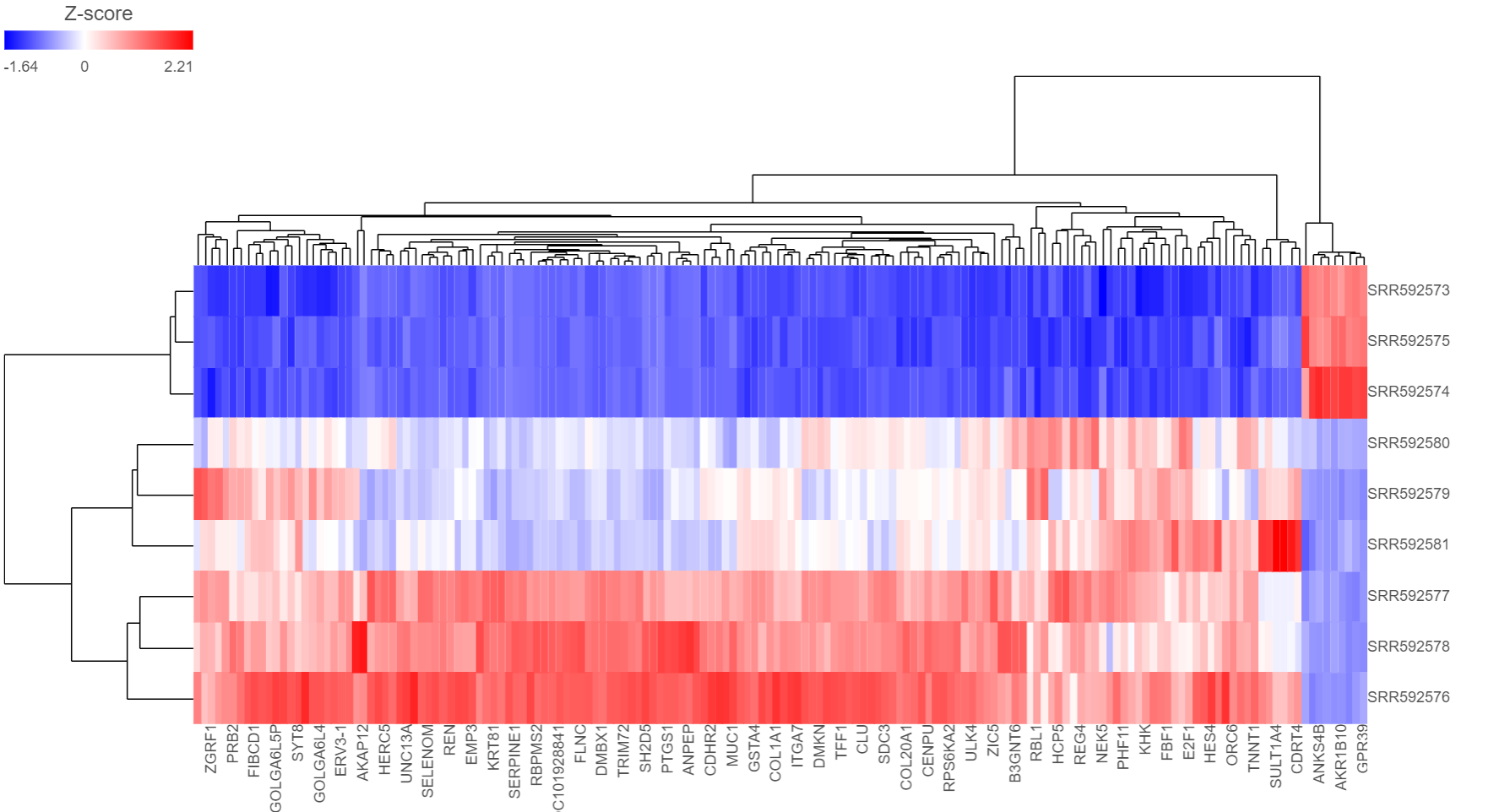
The output of a Hierarchical clustering task is can be a heat map heatmap (Figure 46) or a bubble map with or without dendrogram depends dendrograms depending on whether you perform cluster performed clustering on cells/samples/cells groups or features. By default, samples are on rows (sample labels are displayed as seen in the Data tab) and features (genes or transcripts, depending on the input data) are on columns. Colors are based on standardized expression values (default selection; performed on the fly), with blue indicating low and red indicating high levels of a variable (i. e. low or high expression levels). Dendrograms show clustering of rows (samples) and columns (variables).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Depending on the resolution of your screen and the number of samples and variables (features) that need to be displayed, some binning may be involved. If there are more than samples/genes than pixels, they values of neighboring rows/columns will be averaged together. Use the mouse wheel to zoom in and out. When you zoom in to certain level on the heatmap, you will see each cell represent one sample/gene. To zoom, use the mouse wheel to zoom in / out. When you mouse over the row dendrogram or label area and zoom, it will only zoom in/out on the rows. The binning on the columns will remain the same. Similarly, when you mouse over the column dendrogram or label area and zoom, it will only zoom in/out on the columns. The binning on the rows will remain the same. To move the map around when zoomed in, press down the right button of the mouse and drag the map.
To transpose the map, i.e. to see samples on columns and genes on rows, select the Transpose view button( ). To flip rows or columns belonging to the same dendrogram branch, select the flip mode () button.
When select a cluster by left-clicking & dragging around the dendrogram line in selection mode (), the export button ( will be enabled. Click on it to export the selected features and samples with the normalized values which the heatmap color represents into a text file.
The heat map can be saved as a .svg image by selecting the Save image icon ( ).The control dialog can be seen on Figure 7. The default filename is Dendrogram view.svg and it is downloaded to the local computer.
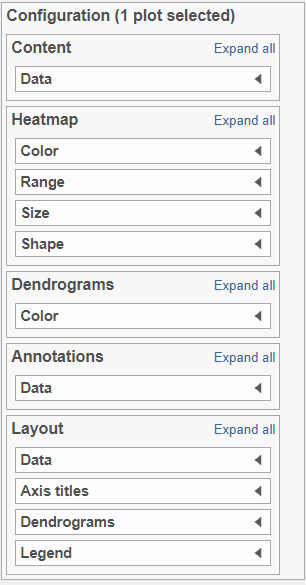
There are 5 sections in the Configuration panel (Figure 5): Content, Heatmap, Dendrograms, Annotations and layout. Click on the triangle () to expand the section to configure.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Content:
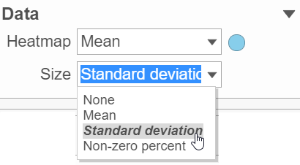
Content contains the value of which matrix data is used to draw heatmap in the plot. Heatmap is a color presentation of the values in the matrix selected. Most of the data nodes contains only one matrix, which you might not need to use Size by configuration to use the same value represent the same information as color by (Figure 6).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
However, if a data node contains multiple matrix information, e.g. if you perform descriptive statistics left mouse button and drag the map. The plot can be saved as a full-size image or as a current view; when Save image is clicked, a prompt will ask how you would like to save the image.
Bubble map
The Hierarchical clustering task can also be used to plot a bubble map. Let's go through the steps to make a bubble map (Figure 7):
- Choose to plot a Bubble map (note the selection of a bubble map in the image which is different from the heatmap). This will open the Bubble map settings.
- Configure the Bubble map settings. First, Group cells by an available categorical attribute (e.g. cell type). Next, summarize the group’s first dimension by color (Group summary method) then choose an additional dimension to plot size (Additional statistic) by using the drop down lists. If these settings are not adjusted, the default dimensions will generate two descriptive statistic measurements that plot the group mean by color and size by the percent of cells. Hierarchical clustering can be performed on the first assigned dimension (by color) which is the Group summary method. The second dimension (size) which is an Additional statistic is not required but it is selected by default (this can be unchecked with the checkbox).
- Ordering the plot columns (Feature order) and rows (Group order) behaves the same as a heatmap. In this example, Ordering for both features and groups by Cluster uses hierarchical clustering to perform distance metrics (default settings will be used but these metrics can be changed under Configure in the Advanced options section). Alternatively, Assign order to features using a managed (saved) feature list or the default order which is dependent on the input data. Assign order to groups can be used to rearrange the attribute by drag and drop, ascending or descending order, or default order which is how the labels as displayed in the Data tab.
- Filtering can be applied to the groups by checking Filter cells then specifying the logical operations to filter by (this is the same as a heatmap).
- Advanced options let the user perform Feature scaling (e.g. Standardize by a z-score) but in a bubble map the default is set to None. It also allows the user to change the Group clustering and Feature clustering options by altering the Cluster distance metrics and Point distance metrics (similar to a heatmap).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Configuration
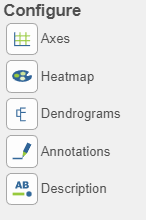
There are plot Configuration/Action options for the Hierarchical clustering / heatmap task which apply to both the heatmap and bubble map in the Data viewer (below): Axes, Heatmap, Dendrograms, Annotations, and Description. Click on the icon to open these configuration options.
Axes
- This section controls the Content or data source used to draw the values in the heatmap or bubble map and also the ability to transpose the axes. The plot is a color representation of the values in the selected matrix. Most of the data nodes contain only one matrix, so it will just say Matrix for the chosen data node. However, if a data node contains multiple matrices (e.g. descriptive statistics were performed on cluster groups for every gene like mean,
...
- standard deviation, percent of
...
- cells, etc
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Heatmap:
Configure the color of the heatmap is
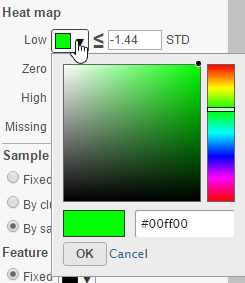
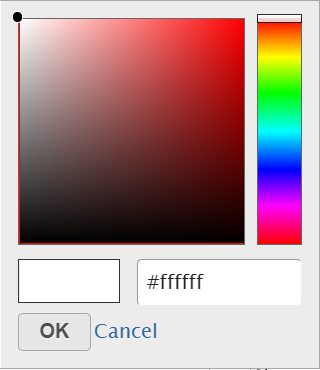
To change a color on the map, click on the arrow head by the color box to get the color mixer (Figure 8). Then pick the color you prefer and select OK. Range of the colors can be changed by typing a different value in the text box to the left of the STD sign (i.e. sandardised).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
- ) each statistic will be in a separate matrix in the output data node. In this case, you can choose which statistic/matrix to display using the drop-down list (this would be the case in a bubble map).
- To change the orientation (switch the columns and rows) of the plot, click on the () toggle switch.
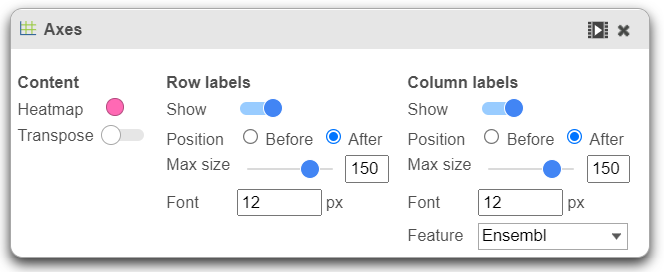
- Row labels and Column labels can be turned on or off by clicking the relevant toggle switches.
- The label size can be changed by specifying the number of pixels using Max size and Font. If an Ensembl annotation model has been associated with the data, you can choose to display the gene name or the Ensembl ID using the Content option.
Heatmap
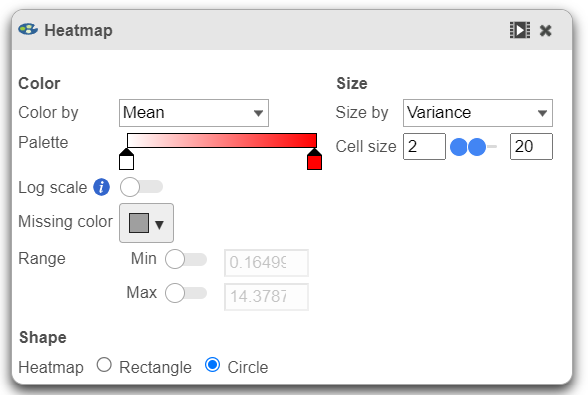
This section is used to configure the color, range, size, and shape of the components in the heatmap.
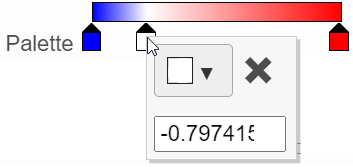
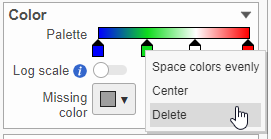
- In the color palette horizontal bar, the left side color represents the lowest value and the right side color represents the highest value in the matrix data. Note that when you zoom in/out the lowest and highest values captured by the color palette may change. By default, there are 3 color stops (): minimum, middle, and maximum color value of the default range calculated on the matrix. Left-click on the middle color stop and drag left or right to change the middle value this color stop represents. If you left-click on the middle color stop once, you can change the color and value this color stop represents (Figure 11). Click on the () to remove this color stop.
- Click on the color square or the adjacent triangle () to choose a color to represent the value. This will display a color picker dialog which allows selection of a color, either by clicking or by typing a HEX color code, then clicking OK.
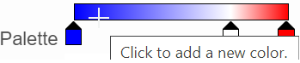
- The min and max color stops cannot be dragged or removed. If you left-click on them, you can choose a different color. When you click on the Palette bar, you can add a new color stop between min and max. Adding a color stop can be useful when there is an outlier value in the data. You can use a different color to represent different value ranges.
- Right-clicking a color stop will reveal a list of options. Space colors evenly will rearrange the position of the stops so there is an equal distance between all stops. Center will move a stop to the middle of the two adjacent stops. Delete will remove the stop.
- To change the min and max threshold values represented by the color palette, click on the toggle switch () under Range, and specify the values in the text boxes.
- In addition to color, you can also use the Size drop-down list to size by a set of values from another matrix stored in the same data node. Most of the data nodes contain only one matrix, so the only options available in the Size drop down will be None or Matrix. In cases where you have multiple matrices, you might want to use the color of the component in the heatmap to represent one type of statistic (like mean of the groups) and the size of the component to represent the information from a different statistic (like std. dev).
- The shape of the heatmap cell (component) can be configured either as a rectangle or circle by selecting the radio button under Shape.
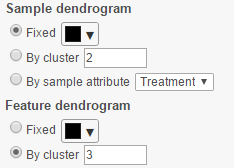
Dendrograms
If cluster analysis is performed on samples and/or features, the result will be displayed as dendrograms. By default, the dendrograms are all colored in black (Figure 9 ).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
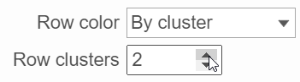
When the By cluster radio button is chosen,
The color of the dendrograms can be configured.
- Click on the color square or its triangle () to choose a different color for the dendrogram.
- When the By cluster in the Row/Column color drop-down list, the number of clusters needs to be specified
...
- . The top N
...
- clusters will be in N different colors.
...
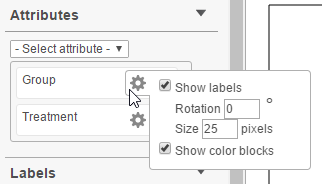
Most of the times you will want to see sample attributes on the heat map. Select the Select attribute drop down list and point to the attribute (as defined in the Data tab) that you want to add to the dendrogram. To remove an attribute from the dendrogram, select the red minus icon ( ). Selecting the gear icon ( ) opens the configuration dialog (Figure 10), which is used to manipulate the dendrogram annotation. Group labels can be turned on or off (Show labels), rotated (Rotation) or re-sized (Size). The color blocks around the labels can also be turned on or off (Show color blocks).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
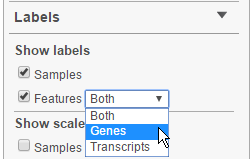
The Labels section of the control panel deals with different labels. First, Show labels turns sample and feature labels on or off. For features with both gene- and transcript level-information, click on the drop-down menu to select which you would like to display, by default, both are shown (Figure 11).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
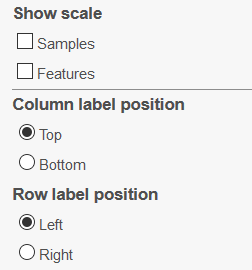
If needed, scales for sample or feature dendrograms can also be added (Show scale). Column labels can be placed at the Top or at the Bottom, while Row labels can be on the Left or on the Right (Figure 12).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
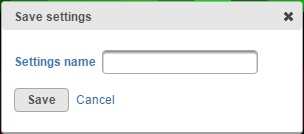
Commonly used plot customization can be saved by selecting the Save settings button. Give your setting set a name (Figure 13) and select Save. Saved settings will be available under the Saved settings list, and can be deleted by clicking on the red cross button( ). To revert to manufacturer settings at any time, use the Default settings hyperlink.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
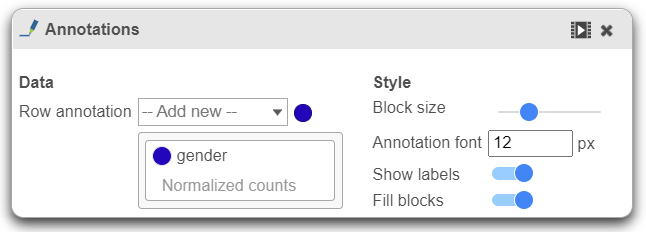
Annotations
This section allows you to add sample or cell level annotations to the viewer. First, make sure to choose the correct data node which contains the annotation information you would like to use by clicking the circle (). All project level annotations will be available on all data nodes in the pipeline.
- Choose an attribute from the Row annotation drop-down list. Multiple attributes can be chosen from the drop-down list and can be reordered by clicking and dragging the groups below the drop-down list. Each attribute is represented as an annotation bar next to the heatmap. Different colors represent the different groups in the attribute.
- The width of the annotation bar can be changed using the Block size slider when the Show labels toggle switch is on.
- The annotation label font size can be changed by specifying the size in pixels.
- The Fill blocks toggle switch adds or removes color from the annotation labels.
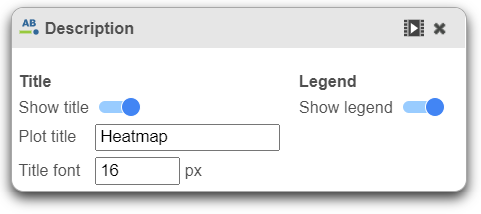
Description
Description is used to modify the Title and toggle on or off the Legend.
In-plot controls
The heatmap has several different mouse modes which modify the way the plot responds to the mouse buttons. The mode buttons are in the upper right corner of the heatmap. Clicking one of these buttons puts the heatmap into that mode.
- In point mode (), you can left-click and drag to move around the heatmap (if you are not fully zoomed out). Left-clicking once on the heatmap or on a dendrogram branch will select the associated rows/columns.
- In selection mode (), you can click and drag to select a range of rows, columns, or components.
- In flip mode (), you can click on a line in the dendrogram (which represents a cluster branch) and the location of the two legs of the branch will be swapped. If no clustering is performed (no dendrogram is generated), in this mode, you can click on the label of an item (observation or feature), drag and drop to manually switch orders of the row or column on the heatmap.
- Click on reset view () to reset to the default
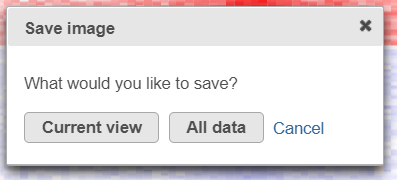
- Save Image icon () enables you to download the heat map to your local computer. If the heat map contains up to 2.5M cells (features * observations), you can choose between saving the current appearance of the heat map window (Current view) and saving the entire heat map (All data). Depending on the number of features / observations, Partek Flow may not be able to fit all the labels on the screen, due to the limit imposed by the screen resolution. All Data option provides an image file of sufficient size so that all the labels are readable (in turn, that image may not fit the compute screen and the image file may be quite large). If the heat map exceeds 2.5M cells, the Current view option will not be shown, and you will see only a dialog like the one below.
- After selecting either Current view (if applicable) or All data button, the next dialog (below) will allow you to specify the image format, size, and resolution.
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...