...
Because we have classified our cells, we can now filter based on those classifications. This can be used to focus on a single cell type for re-clustering and sub-classification or to exclude cells that are not of interest for downstream analysis.
- Click the Classified result Merged counts data node
- Click Filtering
- Click Filter groupscells
- Set to exclude Cell type is Doublets using the drop-down menus
- Click ANDOR
- Set the second filter to exclude Cell type is N/A using the drop-down menus
- Click Finish to apply the filter (Figure 1)
...
| Numbered figure captions |
|---|
| SubtitleText | Set up the Filter groups task to exlcude Doublets and cells that are not classified |
|---|
| AnchorName | Filter groups |
|---|
|
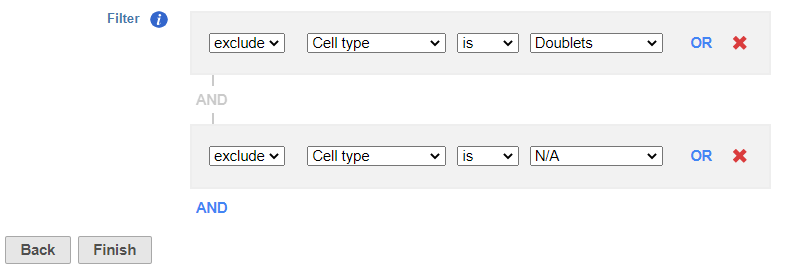 Image Removed Image Removed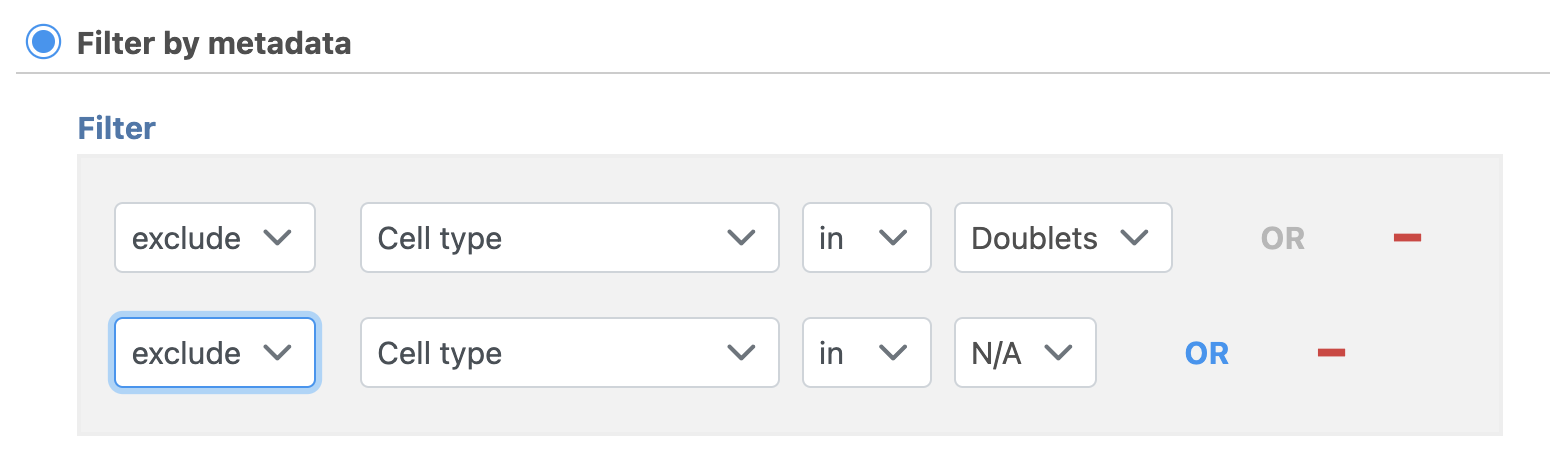 Image Added Image Added
|
This produces a Filtered counts data node (Figure 2).
...
| Numbered figure captions |
|---|
| SubtitleText | Filter groups output |
|---|
| AnchorName | Filtered counts |
|---|
|
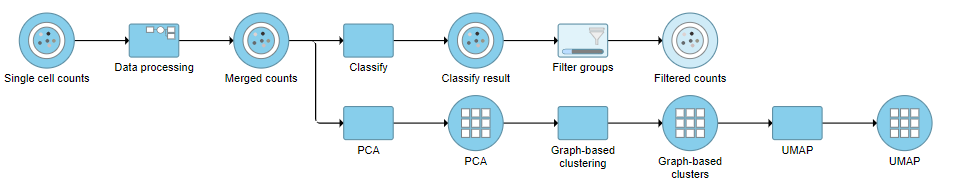 Image Removed Image Removed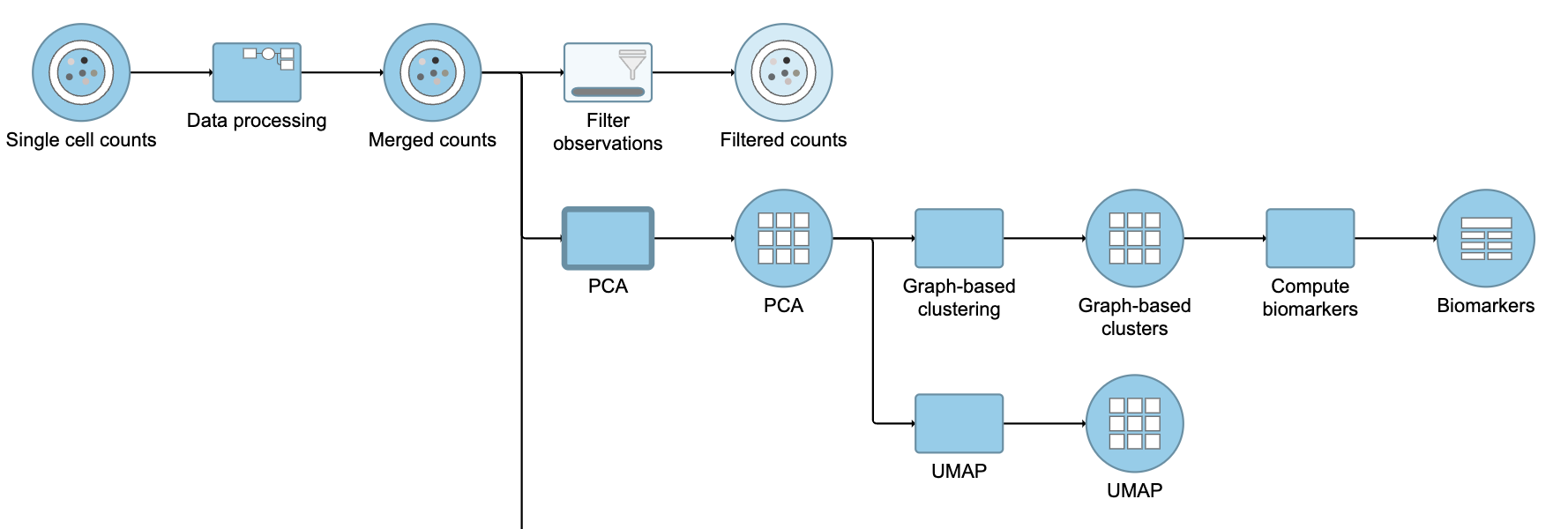 Image Added Image Added
|
Re-split the Matrix
...
| Numbered figure captions |
|---|
| SubtitleText | It is possible to re-split the merged matrix once again |
|---|
| AnchorName | Re-split the matrix |
|---|
|
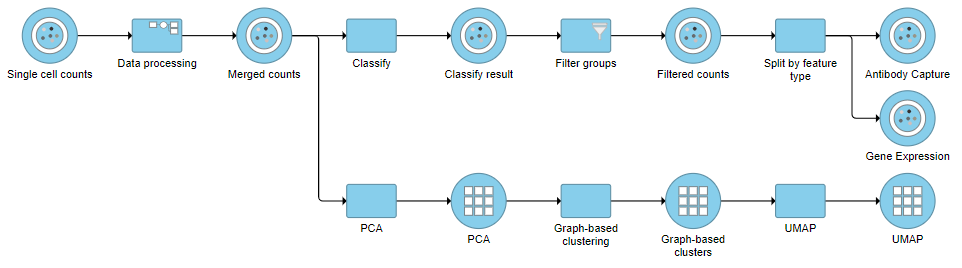 Image Removed Image Removed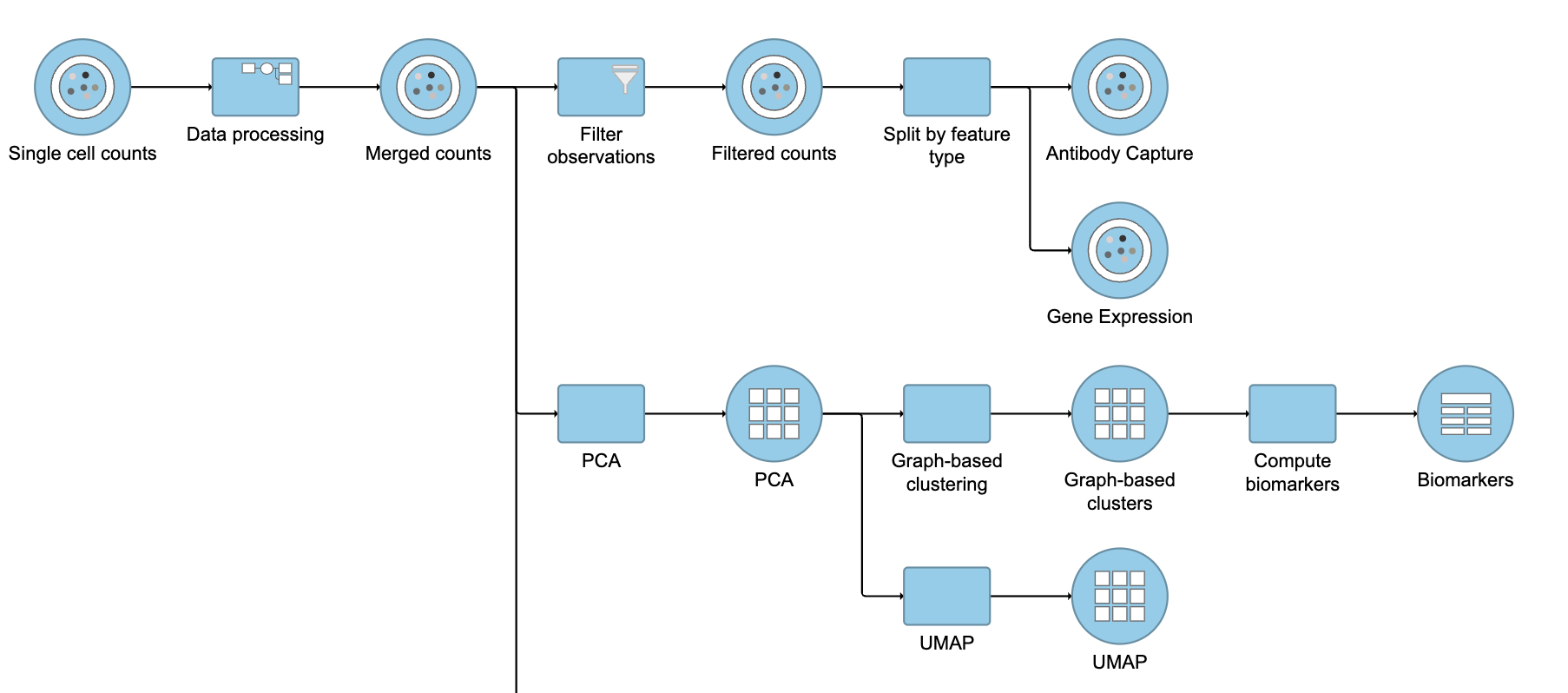 Image Added Image Added
|
Differential Analysis and Visualization - Protein Data
...
- Click the Antibody Capture data node
- Click Statistics
- Click Differential analysis
- Click GSAANOVA then click Next
The first step is to choose which attributes we want to consider in the statistical test.
- Check Click Cell type to include it in the statistical test
- Click Add factor
- Click Next
Next, we will set up the comparison we want to make. Here, we will compare the Activated and Mature B cells.
- Check Drag Activated B cells in the top panel
- Check Drag Mature B cells in the bottom panel
- Click Add comparison
...
| Numbered figure captions |
|---|
| SubtitleText | Setting up a comparison for differentially expressed proteins |
|---|
| AnchorName | CITE-Seq GSA task set up |
|---|
|
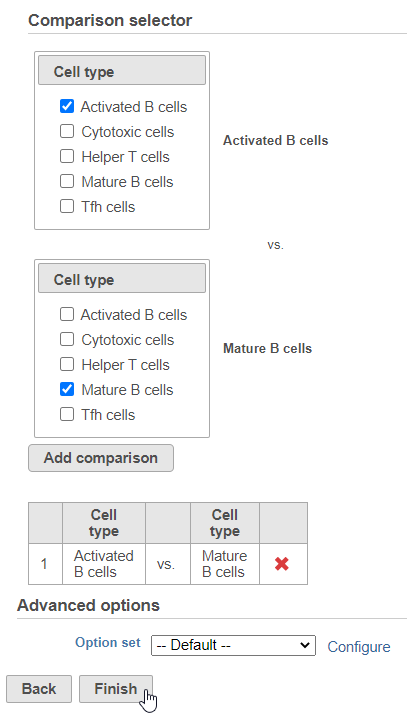 Image Removed Image Removed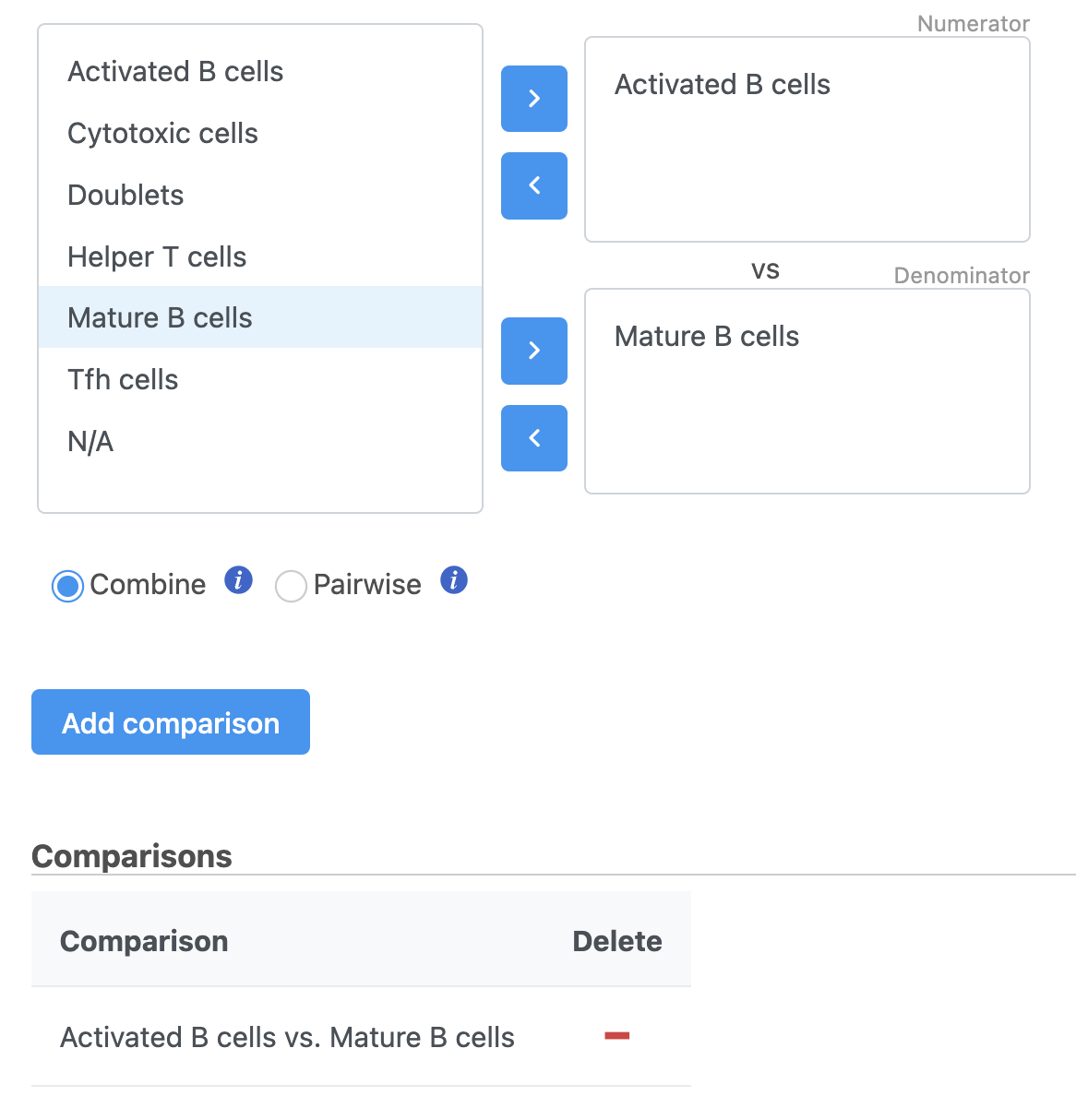 Image Added Image Added
|
The GSA ANOVA task produces a GSA an ANOVA data node.
- Double-click the GSA ANOVA data node to open the task report
...
| Numbered figure captions |
|---|
| SubtitleText | GSA report for protein expression data |
|---|
| AnchorName | GSA protein result |
|---|
|
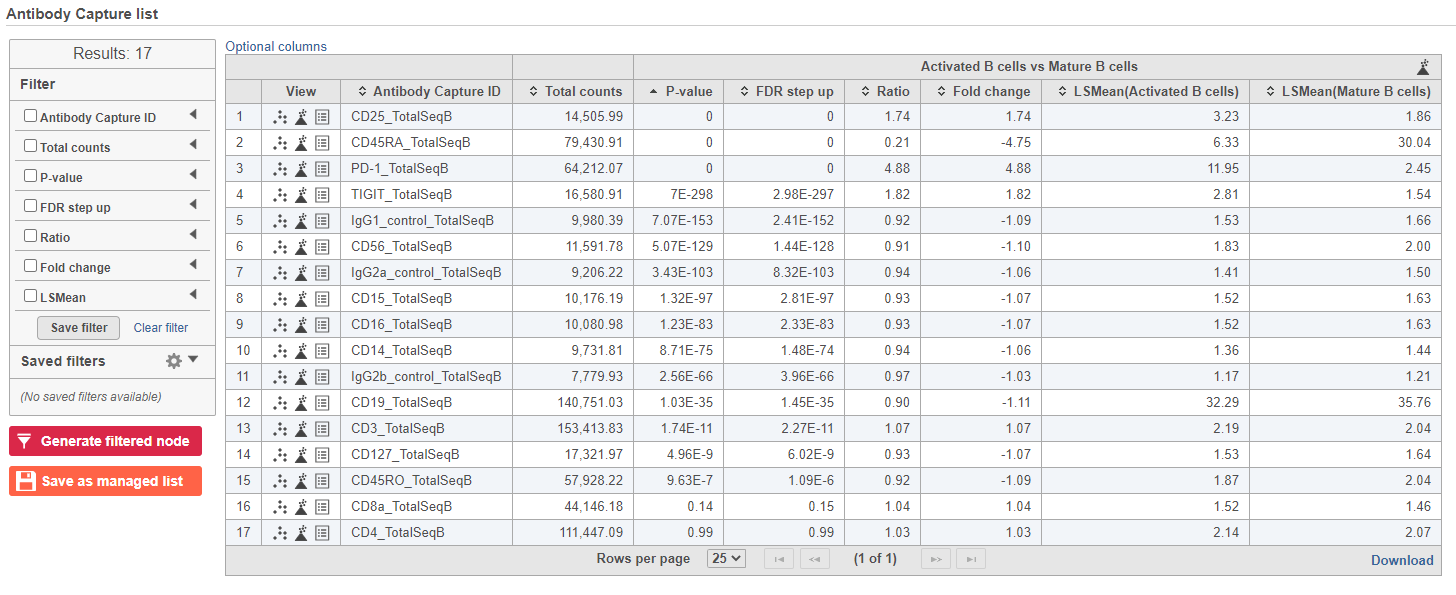 Image Removed Image Removed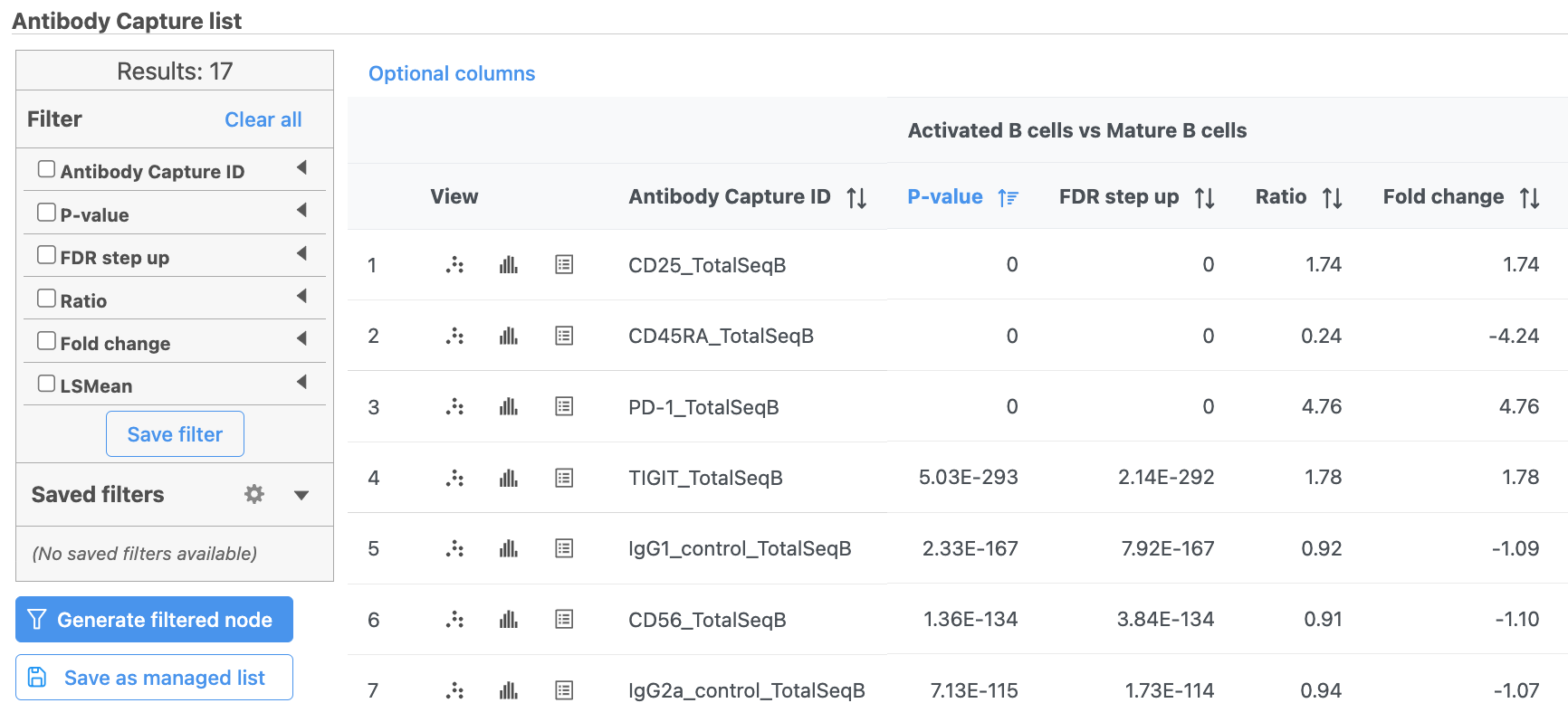 Image Added Image Added
|
In addition to the listed information, we can access dot and violin plots for each gene or protein from this table.
...
This opens a dot plot in a new data viewer session, showing CD45A expression for cells in each of the classifications (Figure 6). First, we exclude Doublets and N/A cells from the plot:
- Open Select and filter, select Criteria
- Drag "Cell type" from the legend title to the Add criteria box
- Uncheck Doublets and N/A
- Click to include selected points
| Numbered figure captions |
|---|
| SubtitleText | CD45RA dot plot for all cells |
|---|
| AnchorName | CD45RA dot plot |
|---|
|
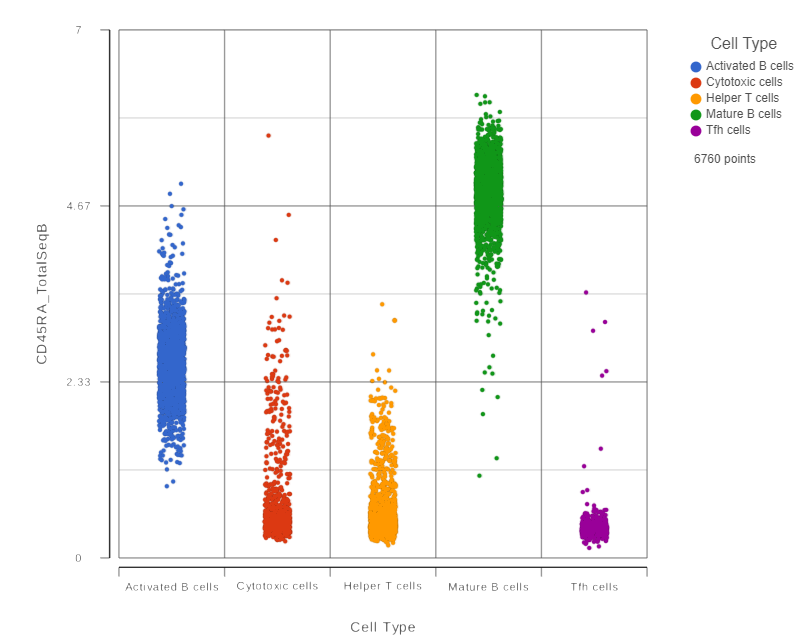 Image Removed Image Removed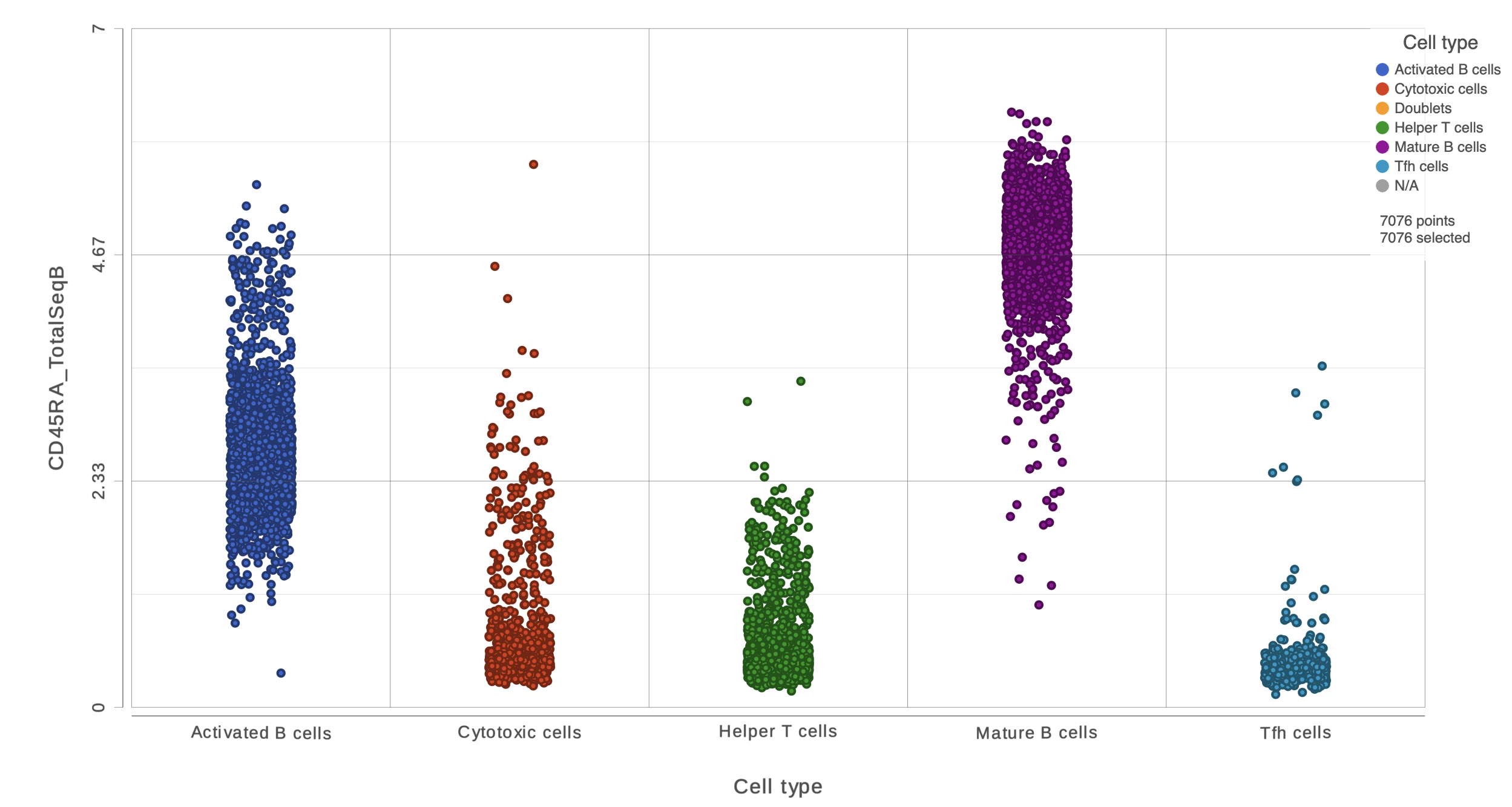 Image Added Image Added
|
We can use the Configuration panel on the left to edit this plot.
- Open the Style icon
- Switch on Violins under Summary
- Switch on Overlay under Summary
- Switch on Colored under Summary
- Select the Graph-based clustering node in the Color by section
- Color by Graph-based clusters under Color and use the slider to decrease the Opacity
- Open the Axes icon
- Select the Graph-based clustering node in the X axis section
- Change the X axis data to Graph-based clusters
- Use the slider to increase the Jitter on the X axis (Figure 7)
...
| Numbered figure captions |
|---|
| SubtitleText | Configure the dot plot using the tools on the left |
|---|
| AnchorName | Configure dot plot |
|---|
|
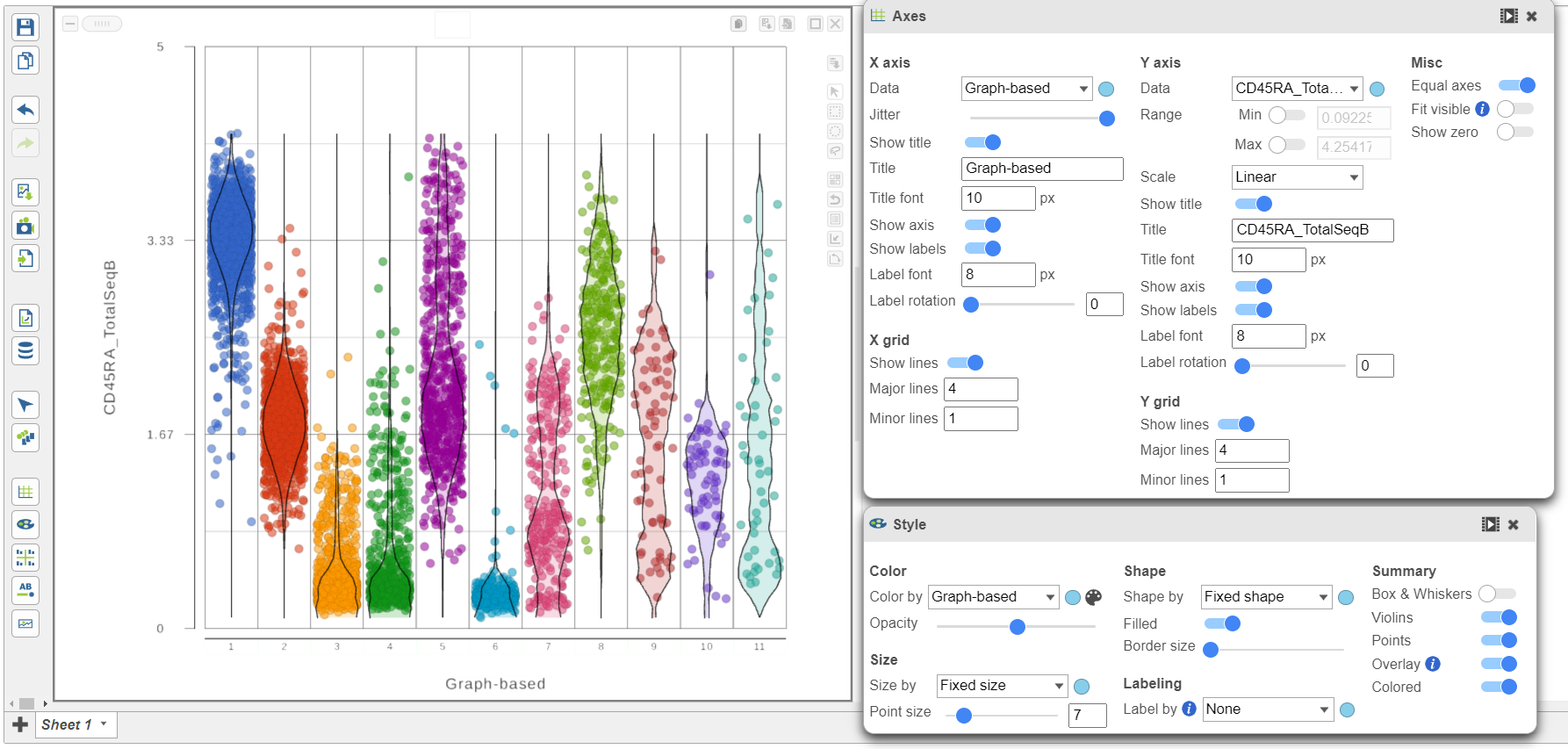 Image Removed Image Removed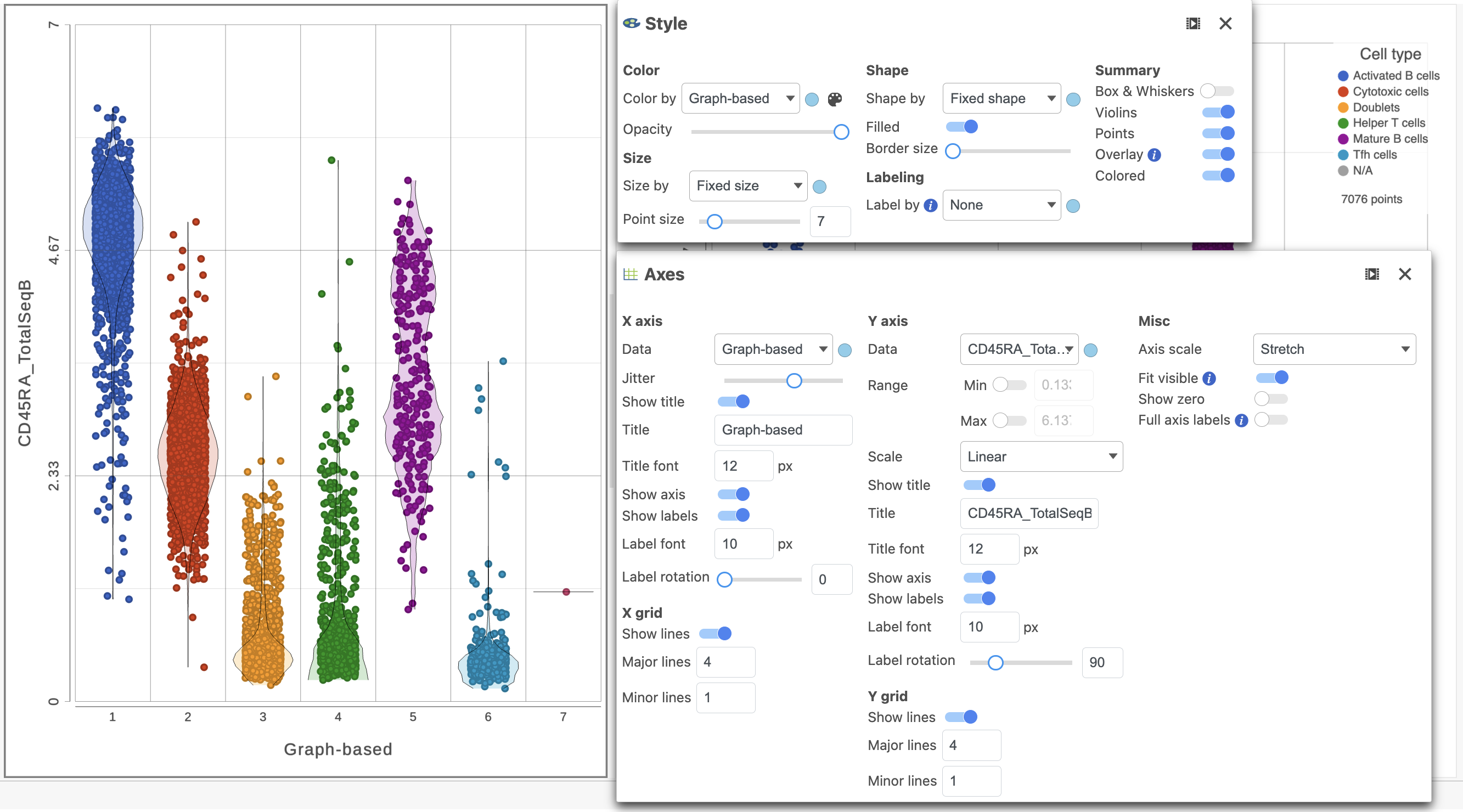 Image Added Image Added
|
- Click the project name to return to the Analyses tab
To visualize all of the proteins at the same time, we can make a hierarchical clustering heat map.
- Click the GSA ANOVA data node
- Click Exploratory analysis in the toolbox
- Click Hierarchical clustering/heatmap
- In the Ordering the Cell order section, choose Graph-based clusters from the Cell Assign order drop-down list
- Click Finish to run with the other default settings
- Double-click the Hierarchical clustering task node to open the heatmap
...
| Numbered figure captions |
|---|
| SubtitleText | Heatmap showing altered Axes labels |
|---|
| AnchorName | Heatmap of proteins |
|---|
|
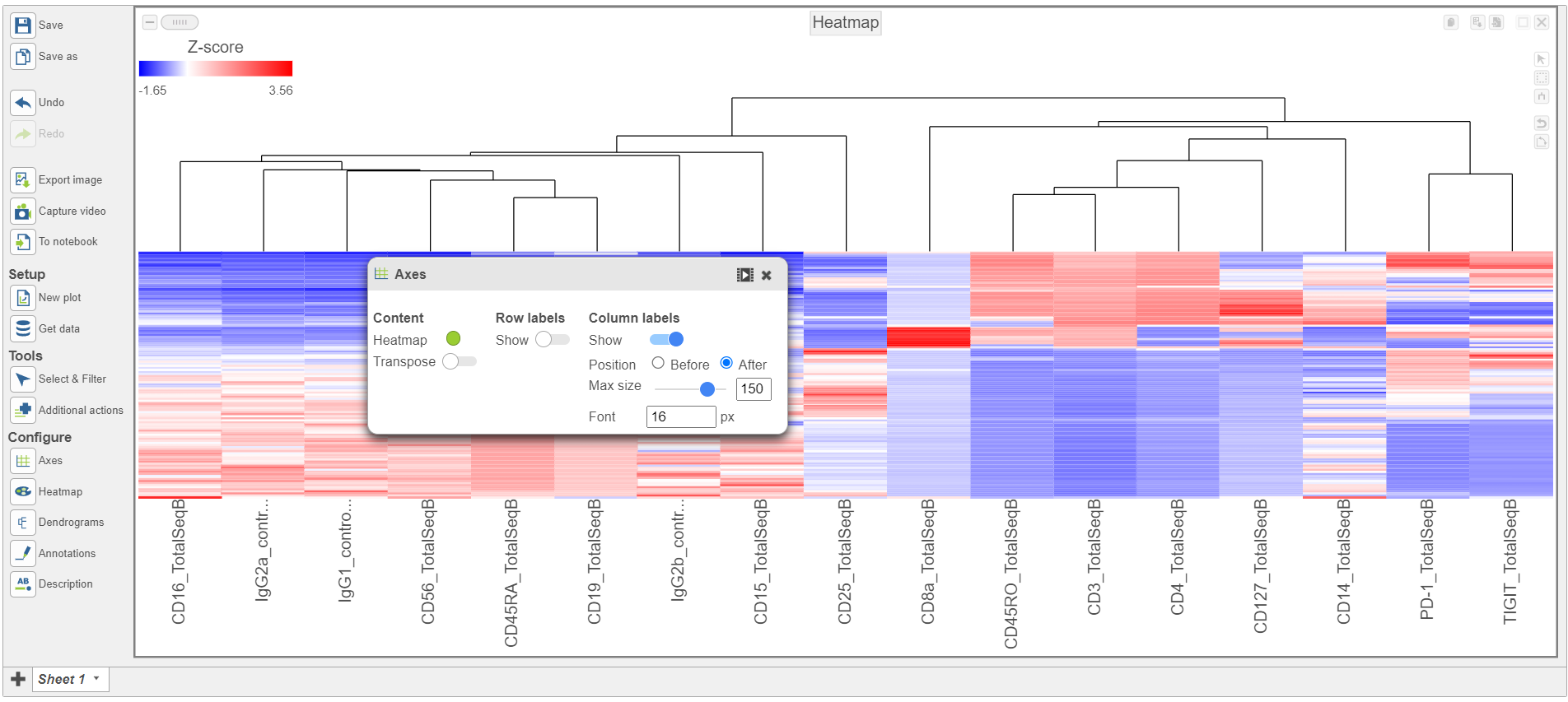 Image Removed Image Removed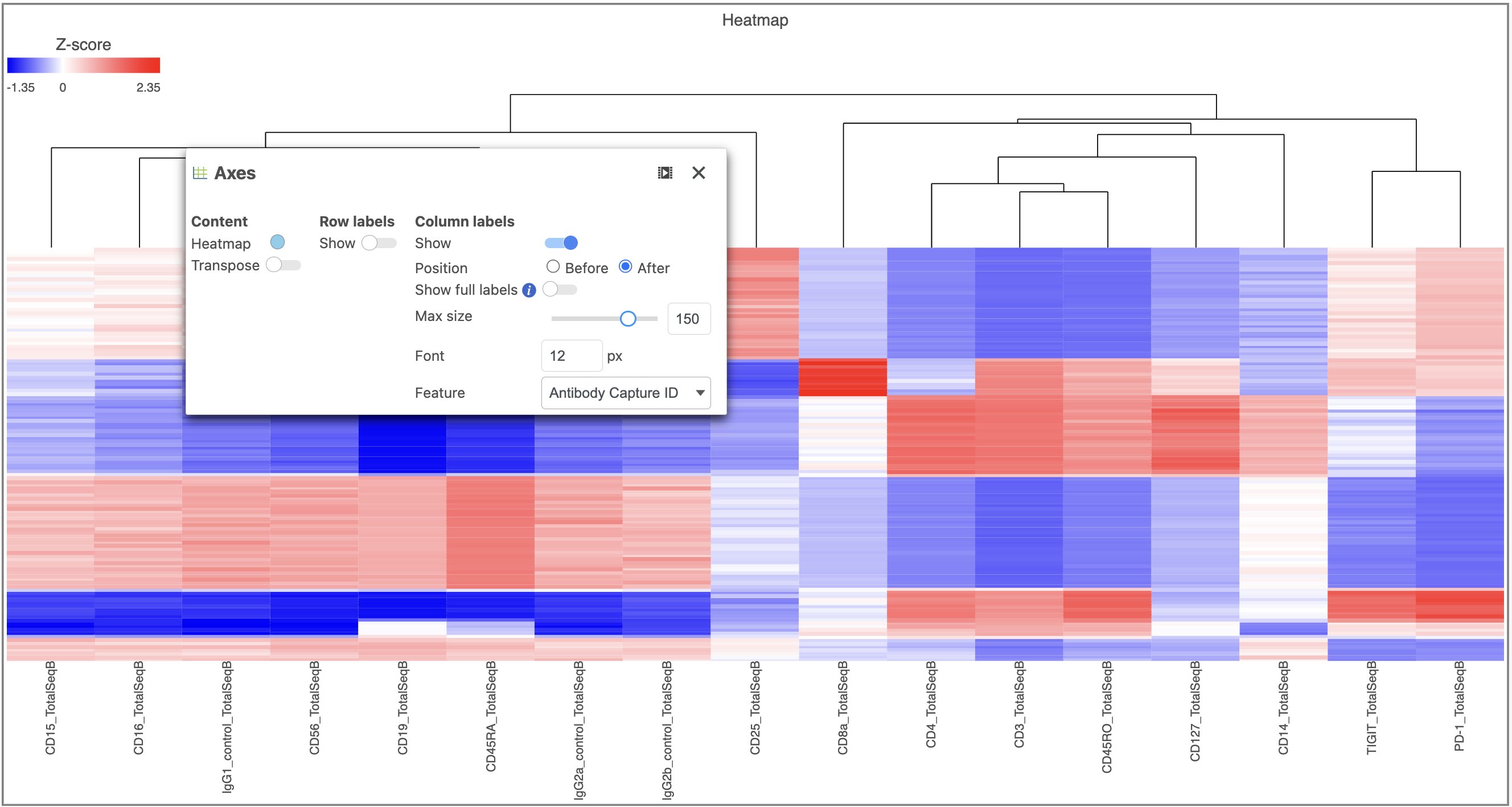 Image Added Image Added
|
- Activate the Transpose switch which will switch the Row and Column labels, so now the Row labels will be shown (Figure 9)
...
| Numbered figure captions |
|---|
| SubtitleText | Transpose the Heatmap to switch the columns and rows |
|---|
| AnchorName | Layout card |
|---|
|
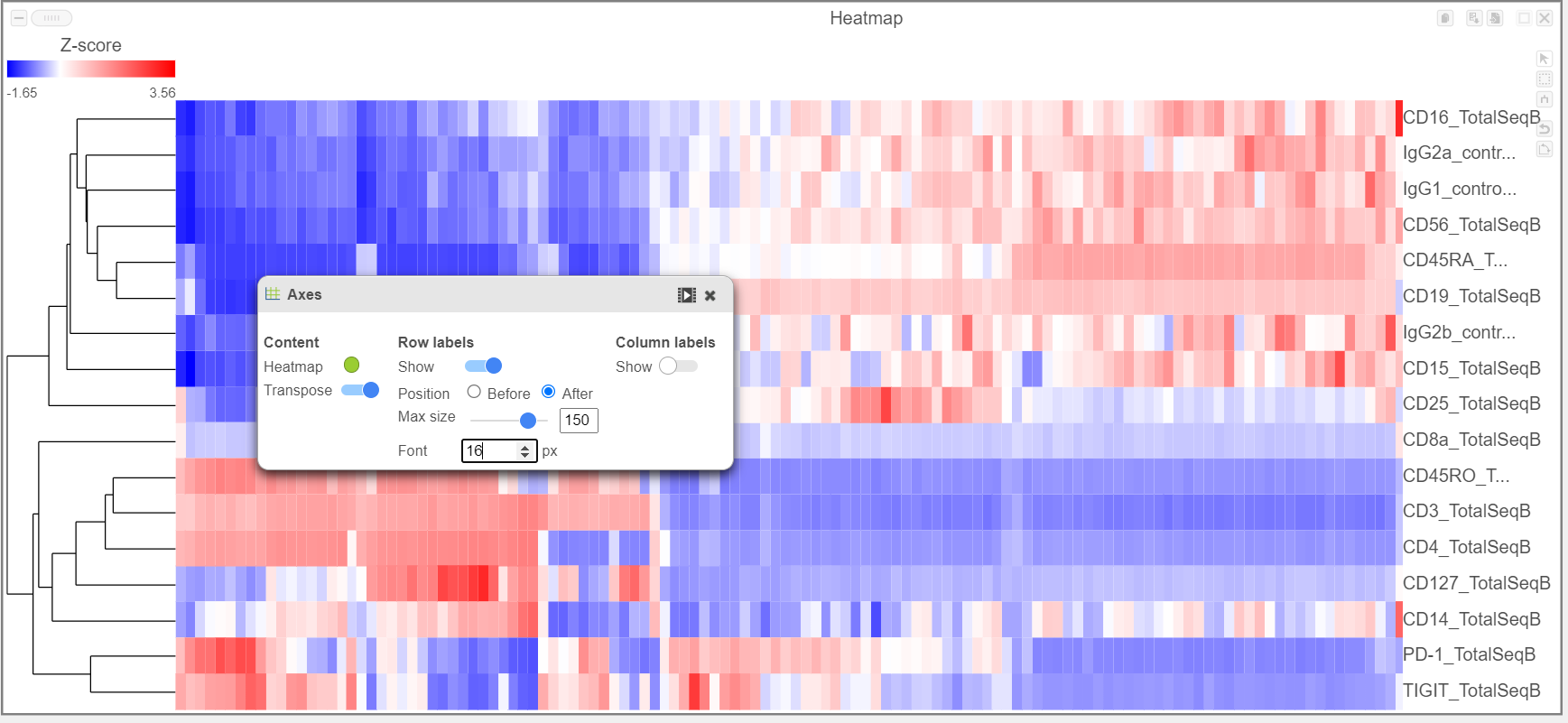 Image Removed Image Removed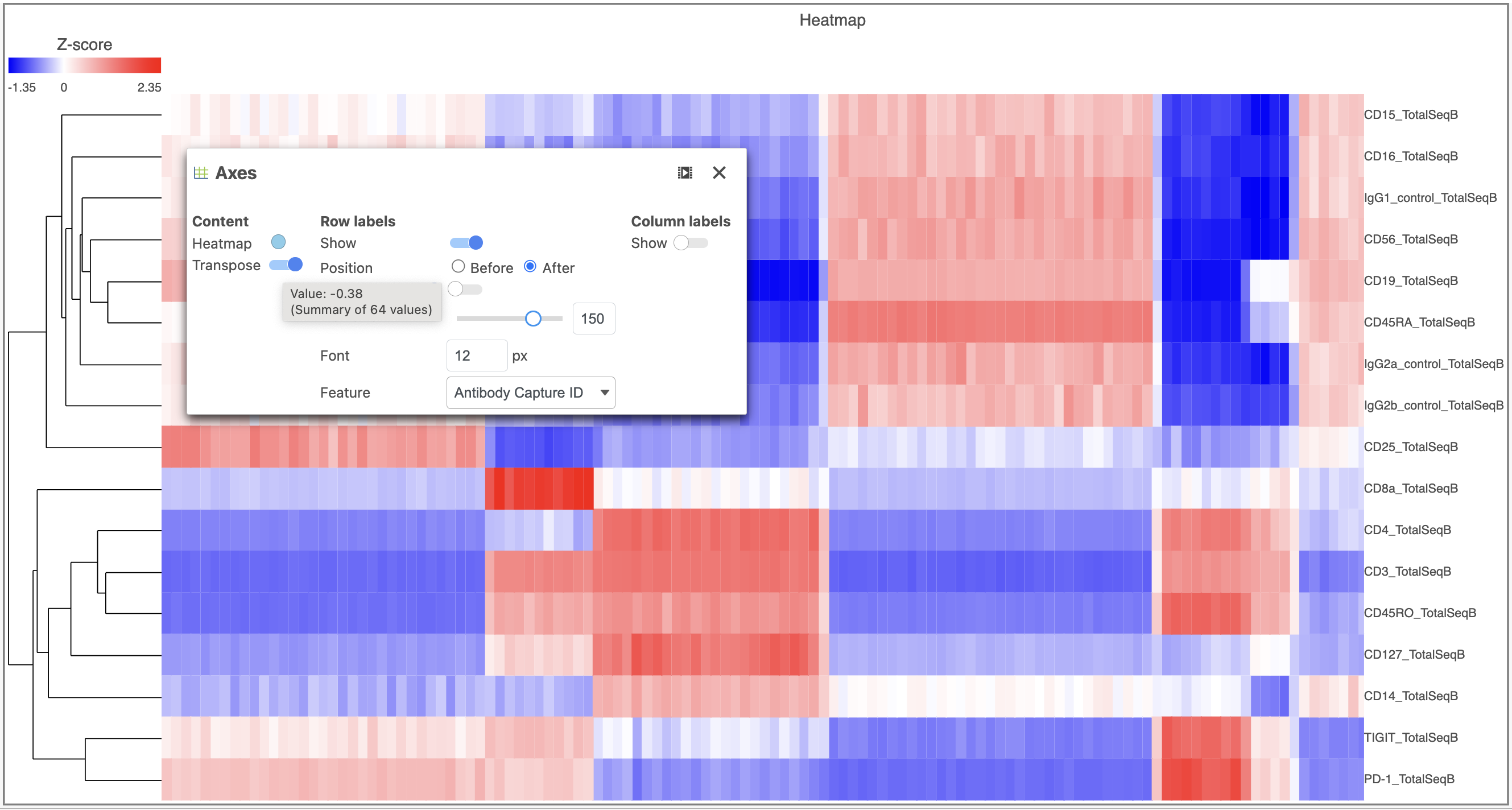 Image Added Image Added
|
- Open the Dendrograms icon
- Choose Row color By cluster and change Row clusters to 4
- Change Row dendrogram size to 80 (Figure 10)
...
| Numbered figure captions |
|---|
| SubtitleText | Configure the Dendrograms settings |
|---|
| AnchorName | Annotations card |
|---|
|
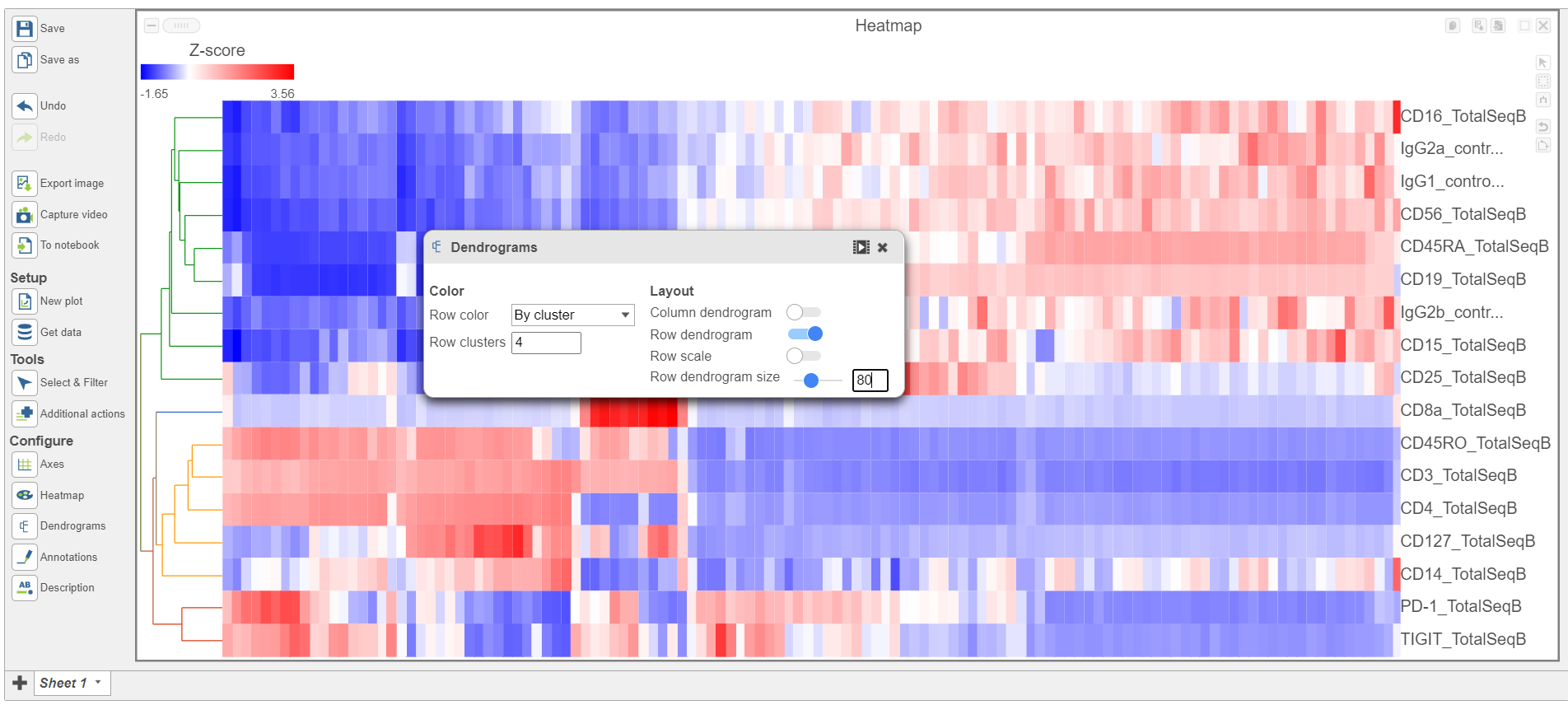 Image Removed Image Removed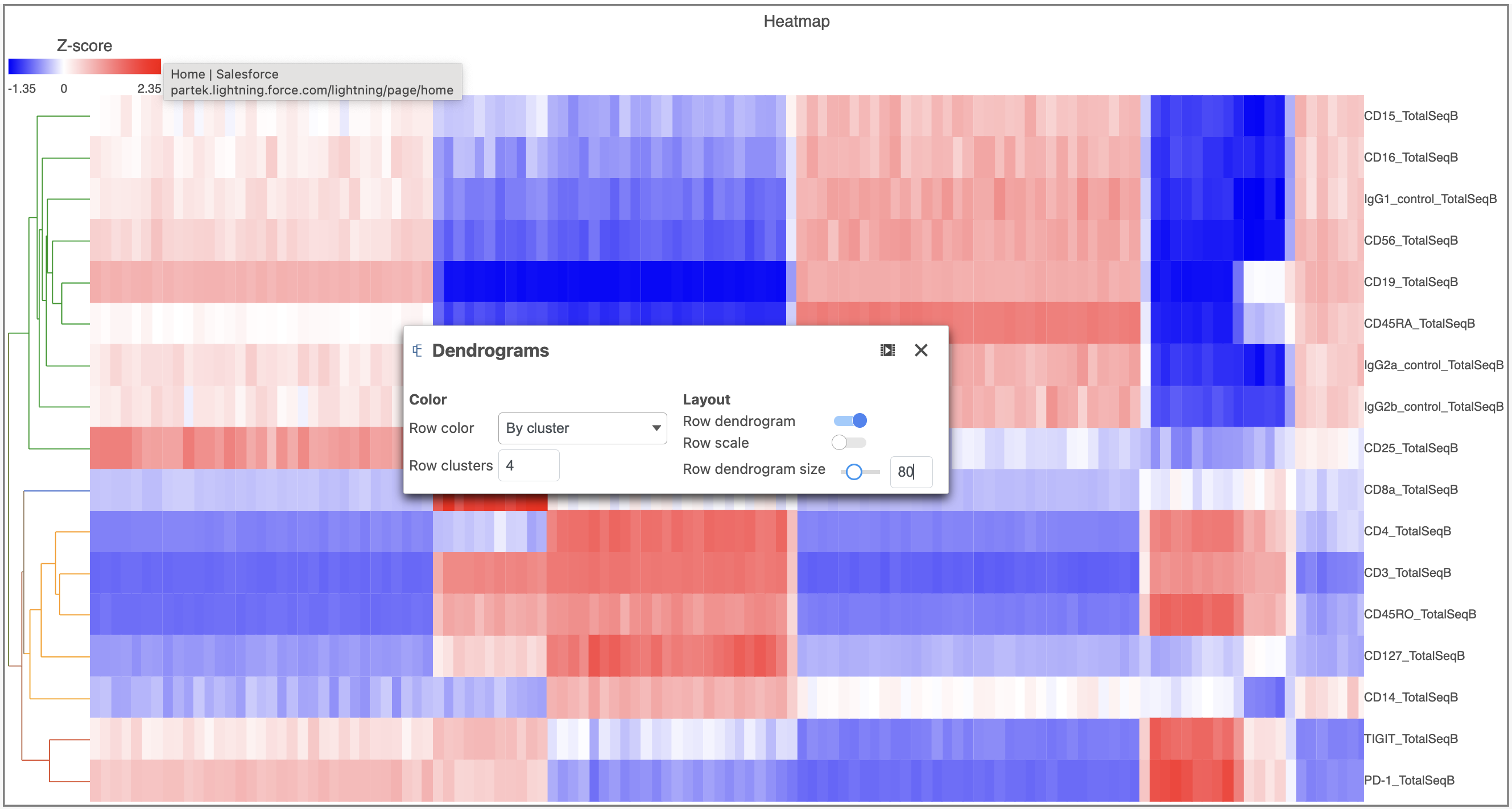 Image Added Image Added
|
- In the Heatmap icon
- Navigate to Range under Color
- Set the Min and Max to -1.2 and 1.2, respectively
- Change the Shape to Circle (Figure 11)
...
| Numbered figure captions |
|---|
| SubtitleText | Configure the Heatmap icon |
|---|
| AnchorName | Heatmap card |
|---|
|
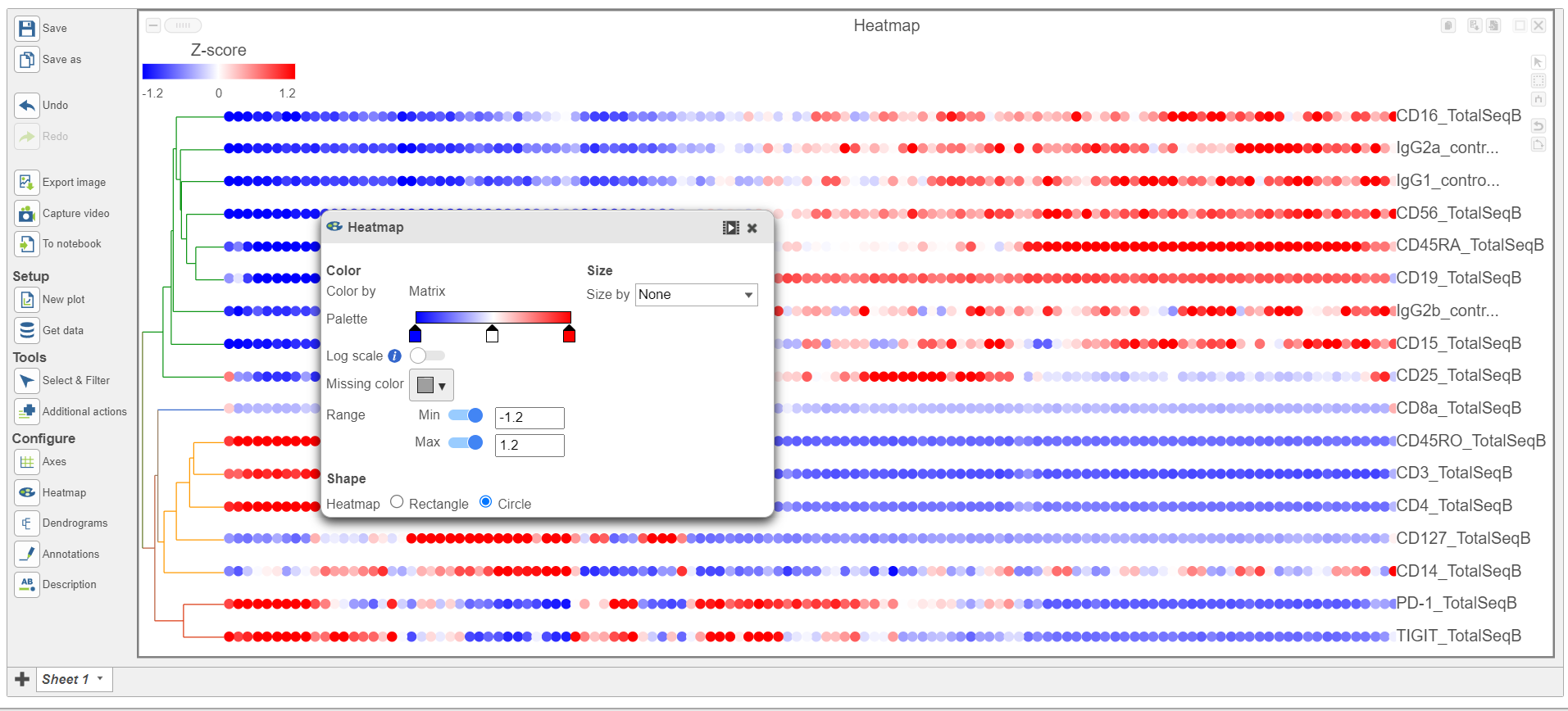 Image Removed Image Removed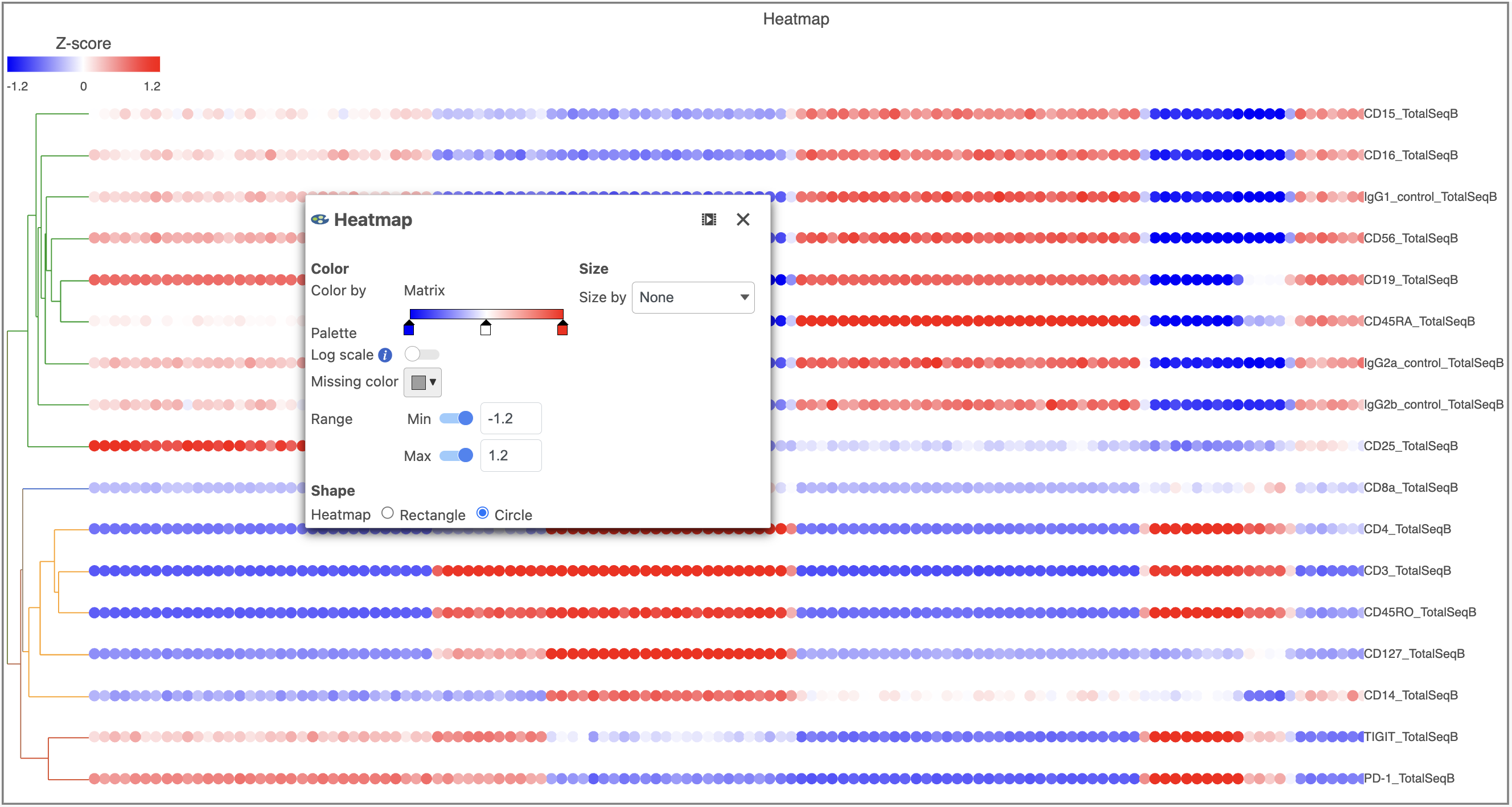 Image Added Image Added
|
- Switch the Shape back to Rectangle
- Change the Color Palette by clicking on the color squares and selecting colors from the rainbow. Click outside of the selection box to exit this selection. The color options can be dragged alone the Palette to highlight value differences (Figure 12).
...
| Numbered figure captions |
|---|
| SubtitleText | Heatmap showing expression of protein markers after changing the Heatmap settings further |
|---|
| AnchorName | Heatmap of proteins configured |
|---|
|
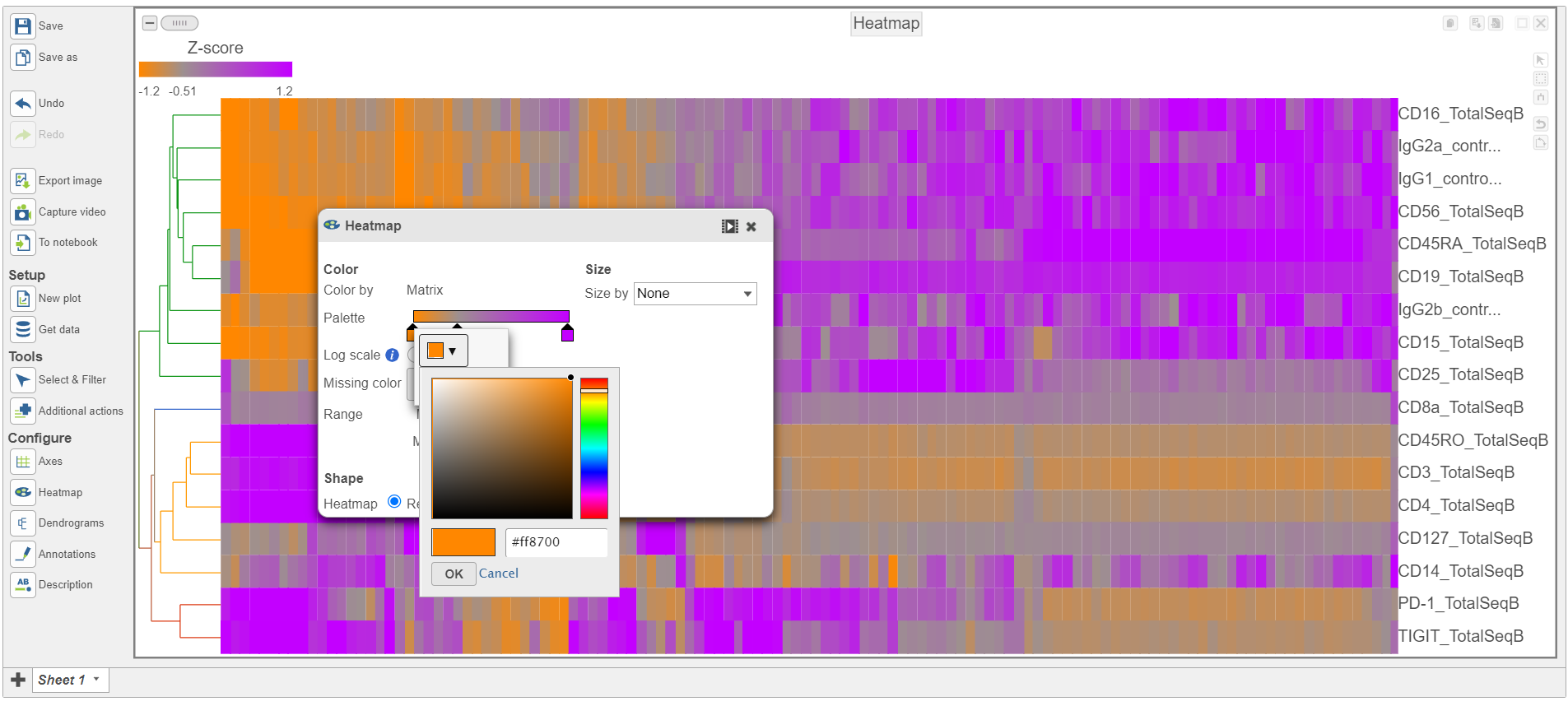 Image Removed Image Removed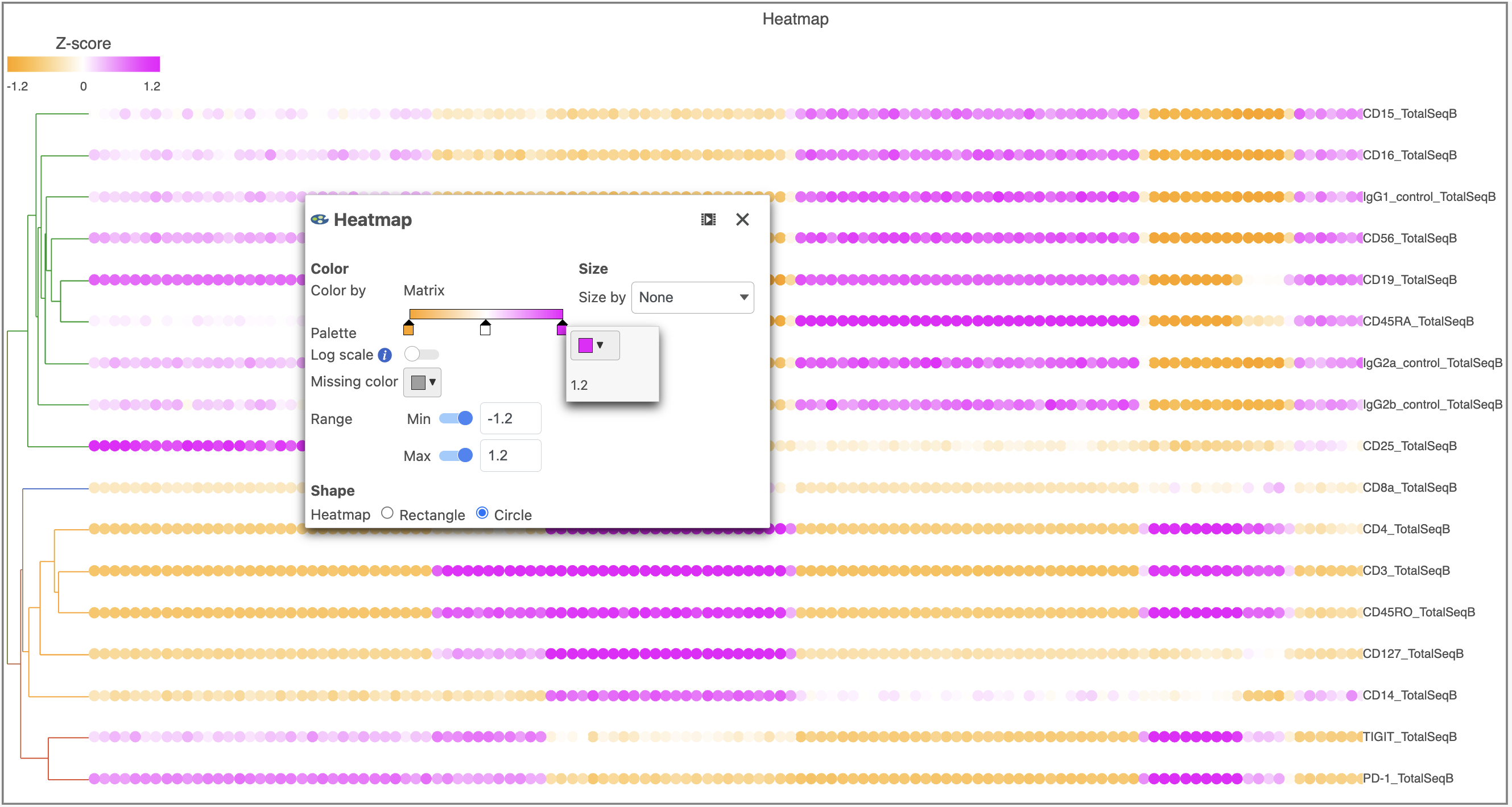 Image Added Image Added
|
Feel free to explore the other tool options on the left to customize the plot further.
...
- Click the project name to return to the Analyses tab
- Click the Gene Expression data node
- Click the Antibody Capture data node
- Click Statistics
- Click Differential analysis
- Click GSA
- Check Cell type to include it in the statistical test
- ANOVA then click Next
- Click Cell type
- Click Add factor
- Click Next
- Check Drag Activated B cells in the top panel
- Check Drag Mature B cells in the bottom panel
- Click Add comparison Click comparison
The comparison should appear in the table as Activated B cells vs. Mature B cells.
- Click Finish to run the statistical test
As before, this will generate a GSA an ANOVA task node and a GSA n ANOVA data node.
- Double-click the GSA ANOVA task node to open the task report (Figure 13)
...
| Numbered figure captions |
|---|
| SubtitleText | GSA report for the gene expression data |
|---|
| AnchorName | GSA genes result |
|---|
|
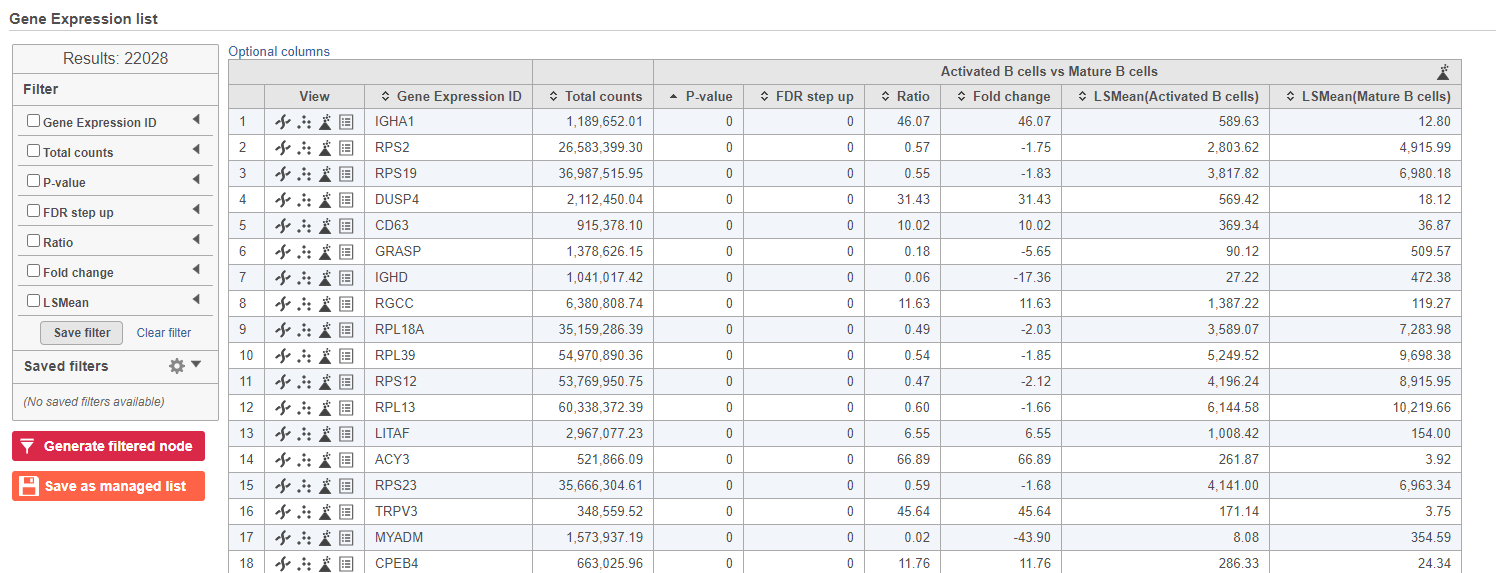 Image Removed Image Removed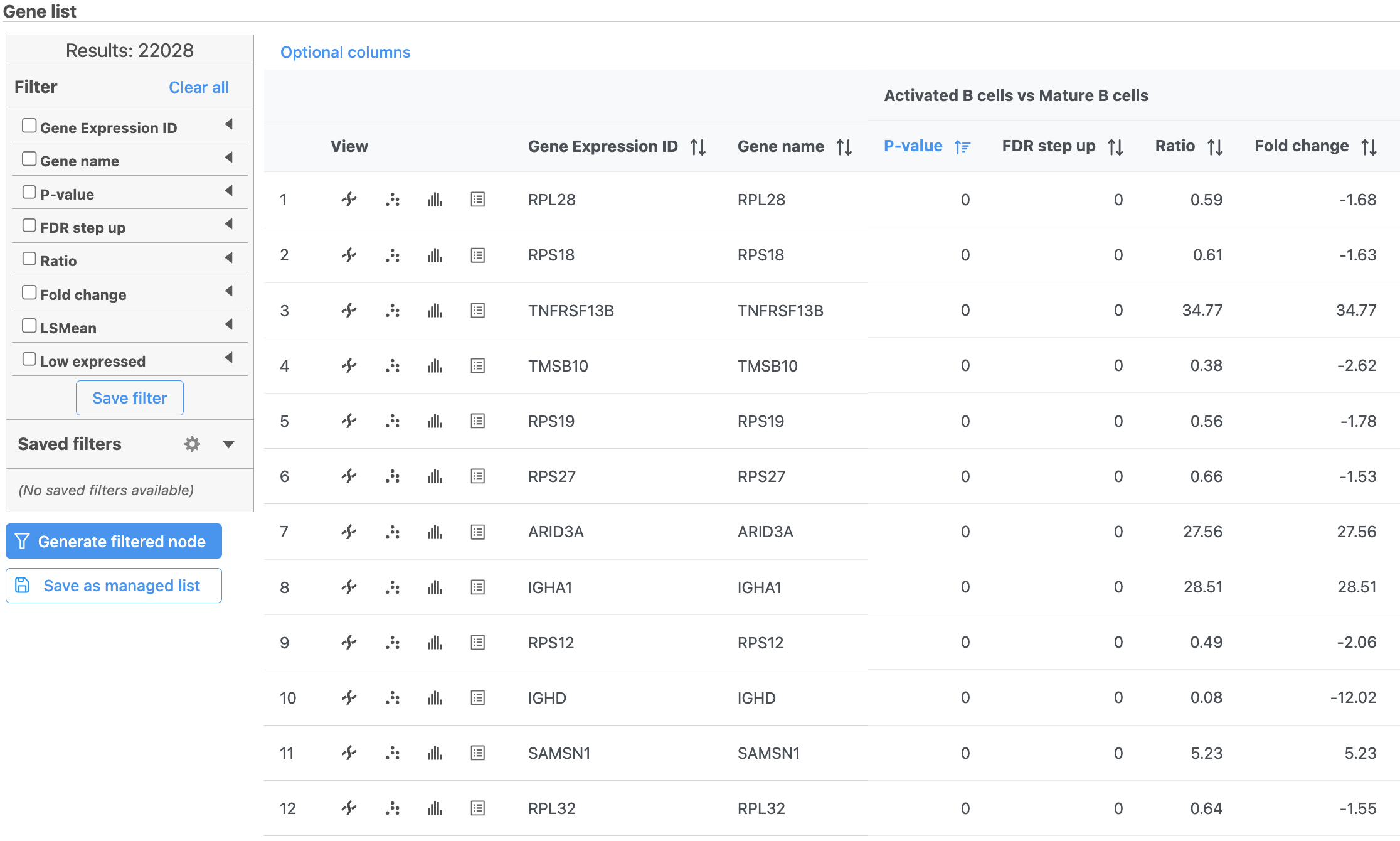 Image Added Image Added
|
Because more than 20,000 genes have been analyzed, it is useful to use a volcano plot to get an idea about the overall changes.
...
| Numbered figure captions |
|---|
| SubtitleText | The volcano plot can be Configured using the icons on the left and in plot controls |
|---|
| AnchorName | Volcano plot gene expression |
|---|
|
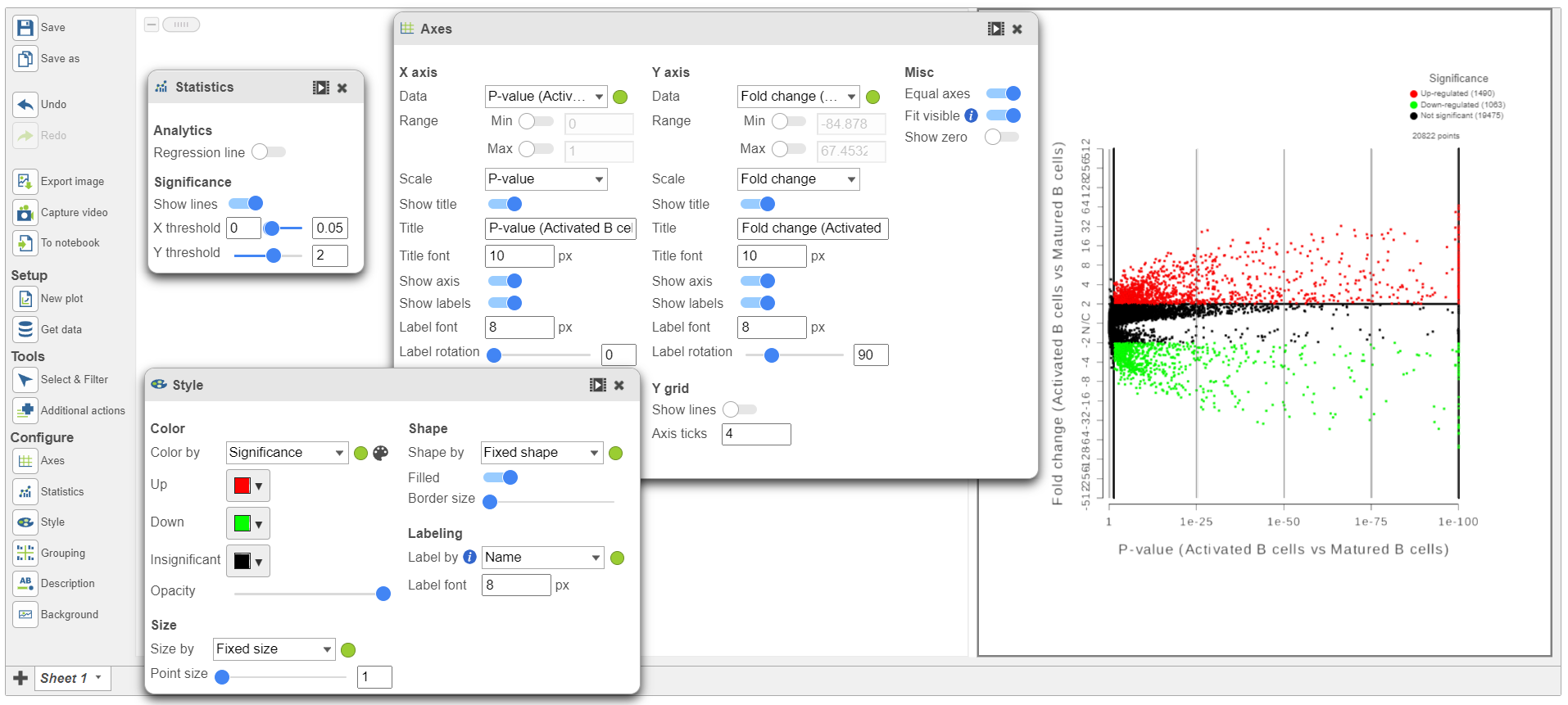 Image Removed Image Removed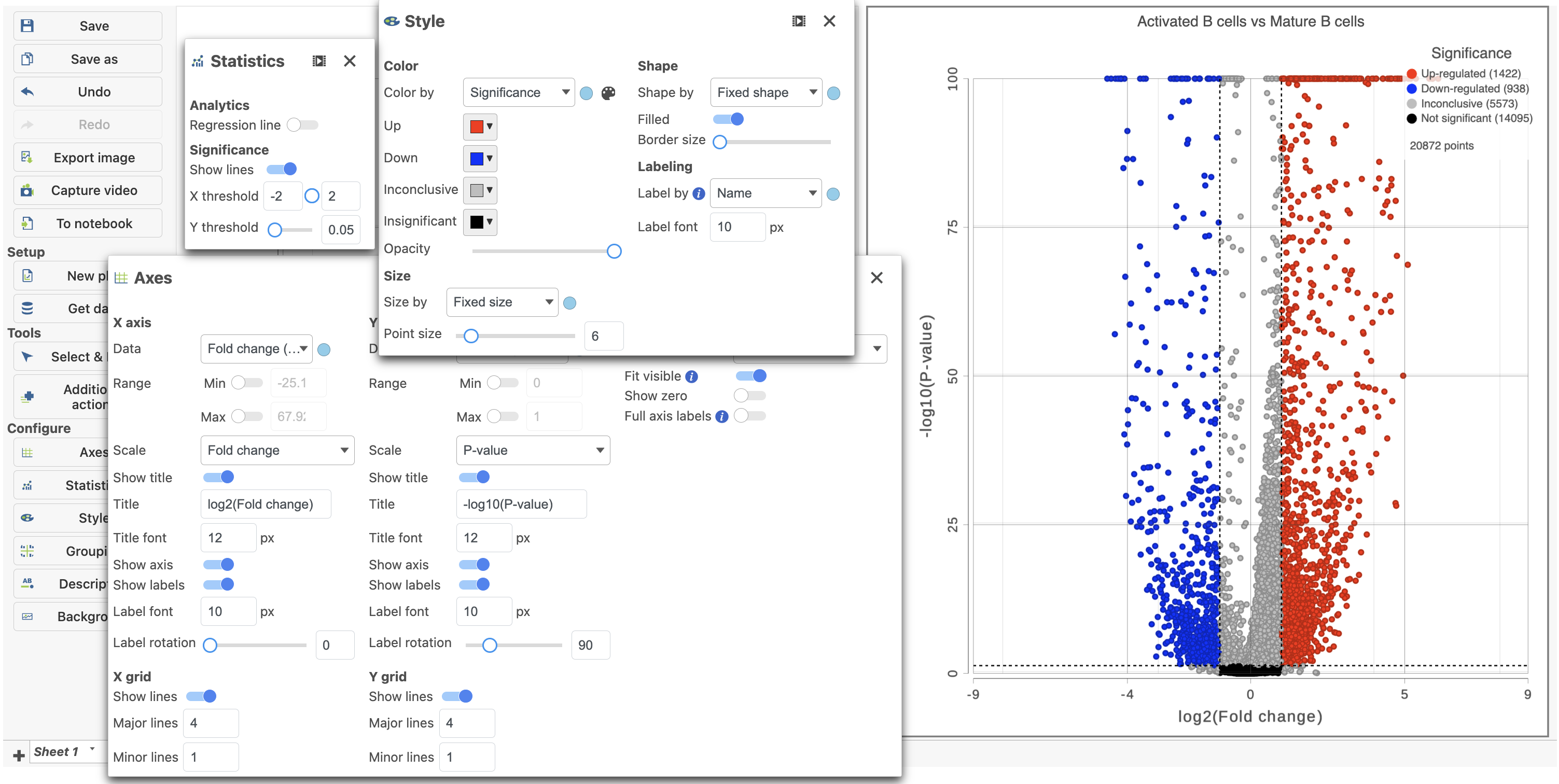 Image Added Image Added
|
- Click the GSA ANOVA report tab in your web browser to return to the full report
...
| Numbered figure captions |
|---|
| SubtitleText | Use the panel on the left to filter the list for significant genes |
|---|
| AnchorName | Significant genes |
|---|
|
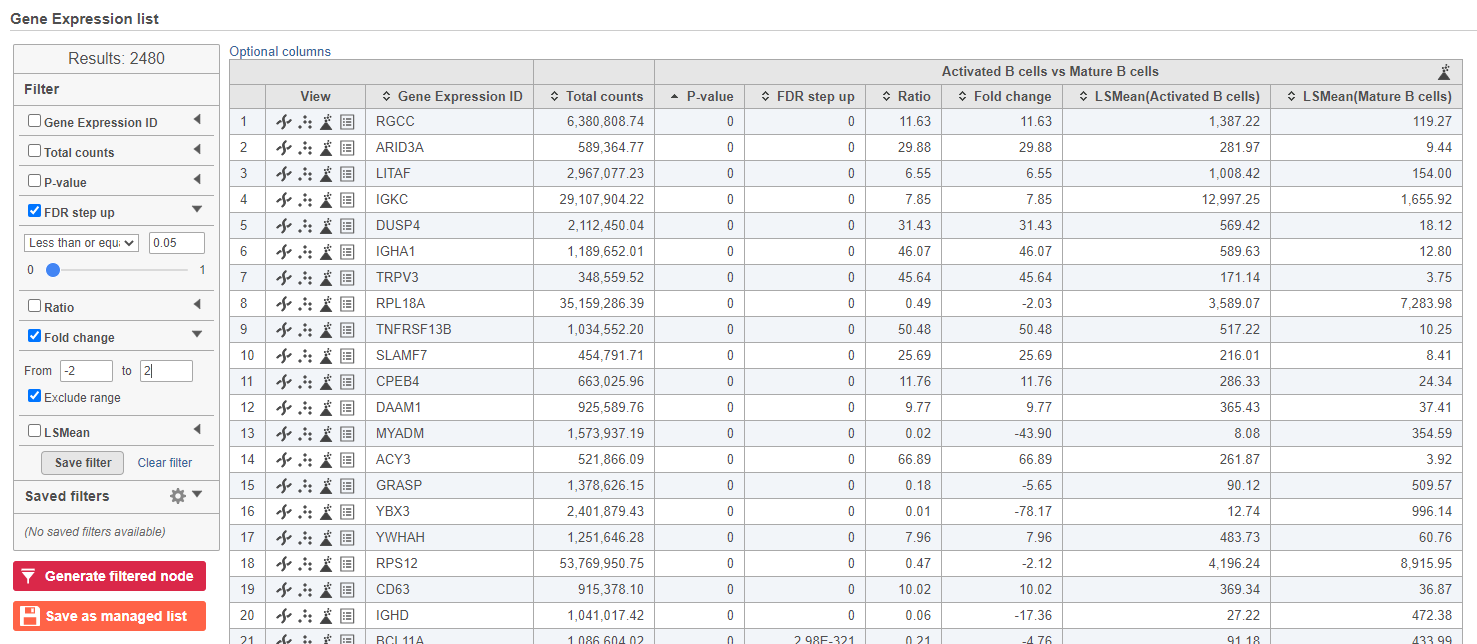 Image Removed Image Removed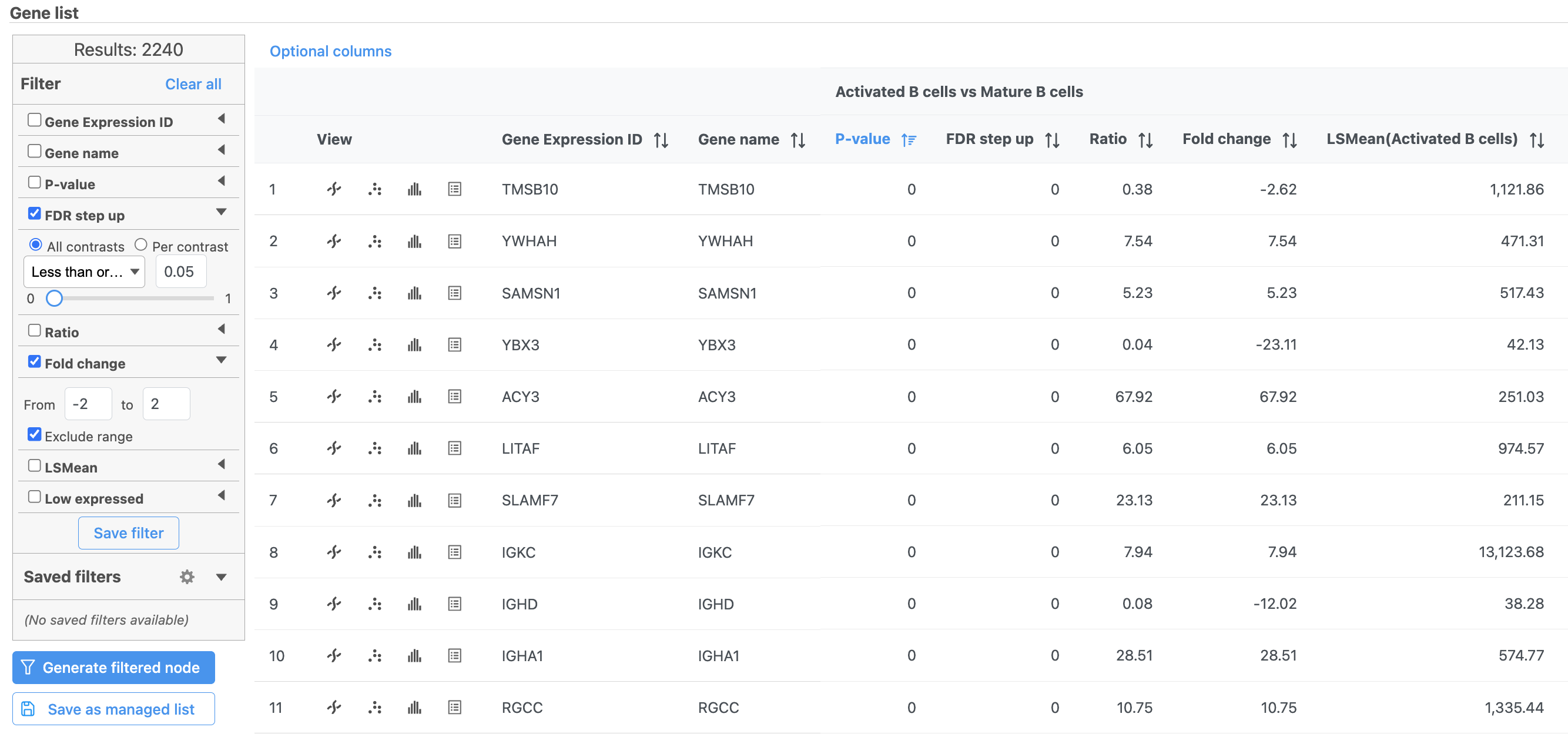 Image Added Image Added
|
- Click
 Image Removed
Image Removed Image Added to create a new data node including only these significantly different genes
Image Added to create a new data node including only these significantly different genes
...
| Numbered figure captions |
|---|
| SubtitleText | Results of pathway enrichment test |
|---|
| AnchorName | Pathway enrichment analysis results |
|---|
|
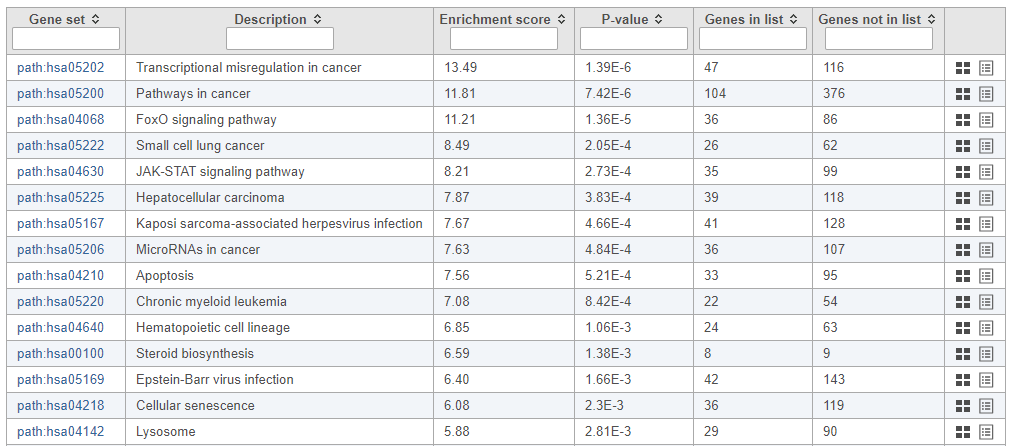 Image Removed Image Removed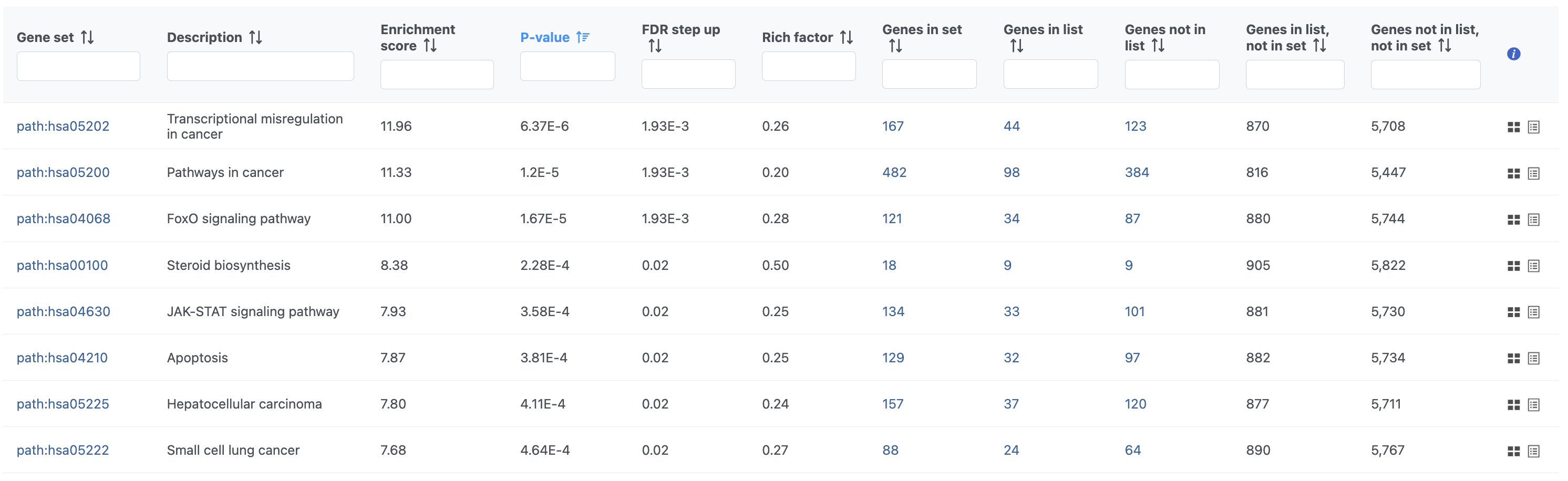 Image Added Image Added
|
To get a better idea about the changes in each enriched pathway, we can view an interactive KEGG pathway map.
...
| Numbered figure captions |
|---|
| SubtitleText | Transcriptional misregulation in cancer pathway with significant genes highlighted in green and red |
|---|
| AnchorName | Transcriptional misregulation in cancer |
|---|
|
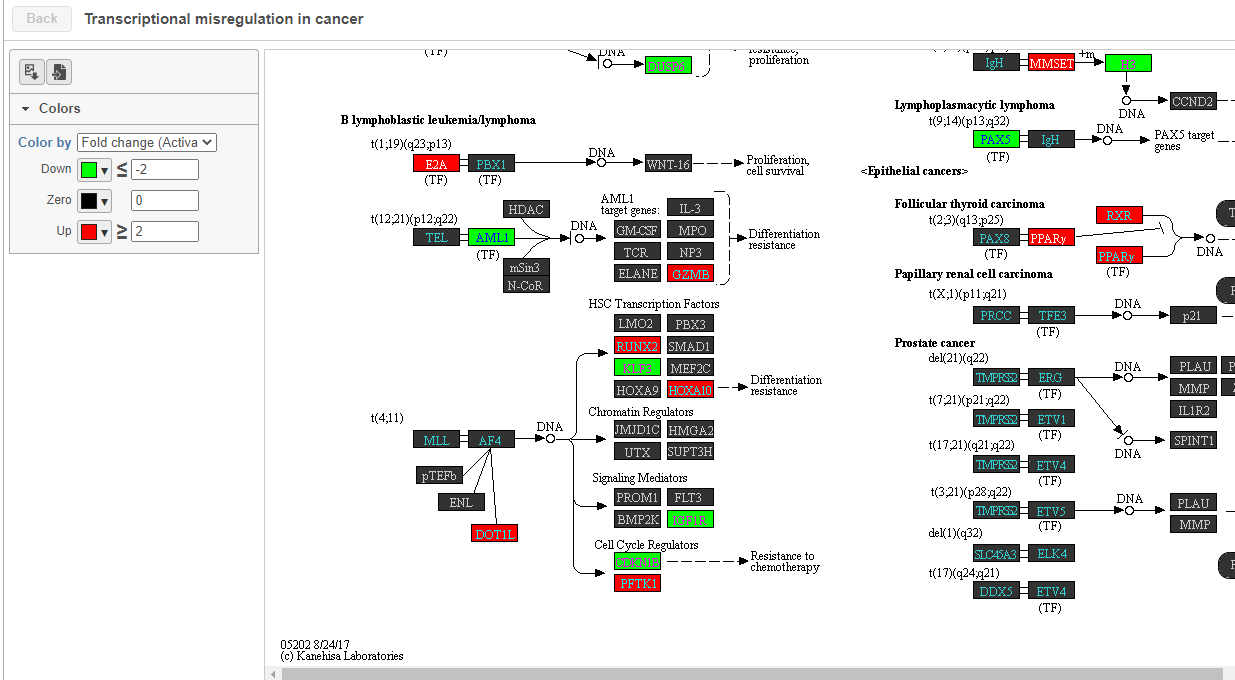 Image Removed Image Removed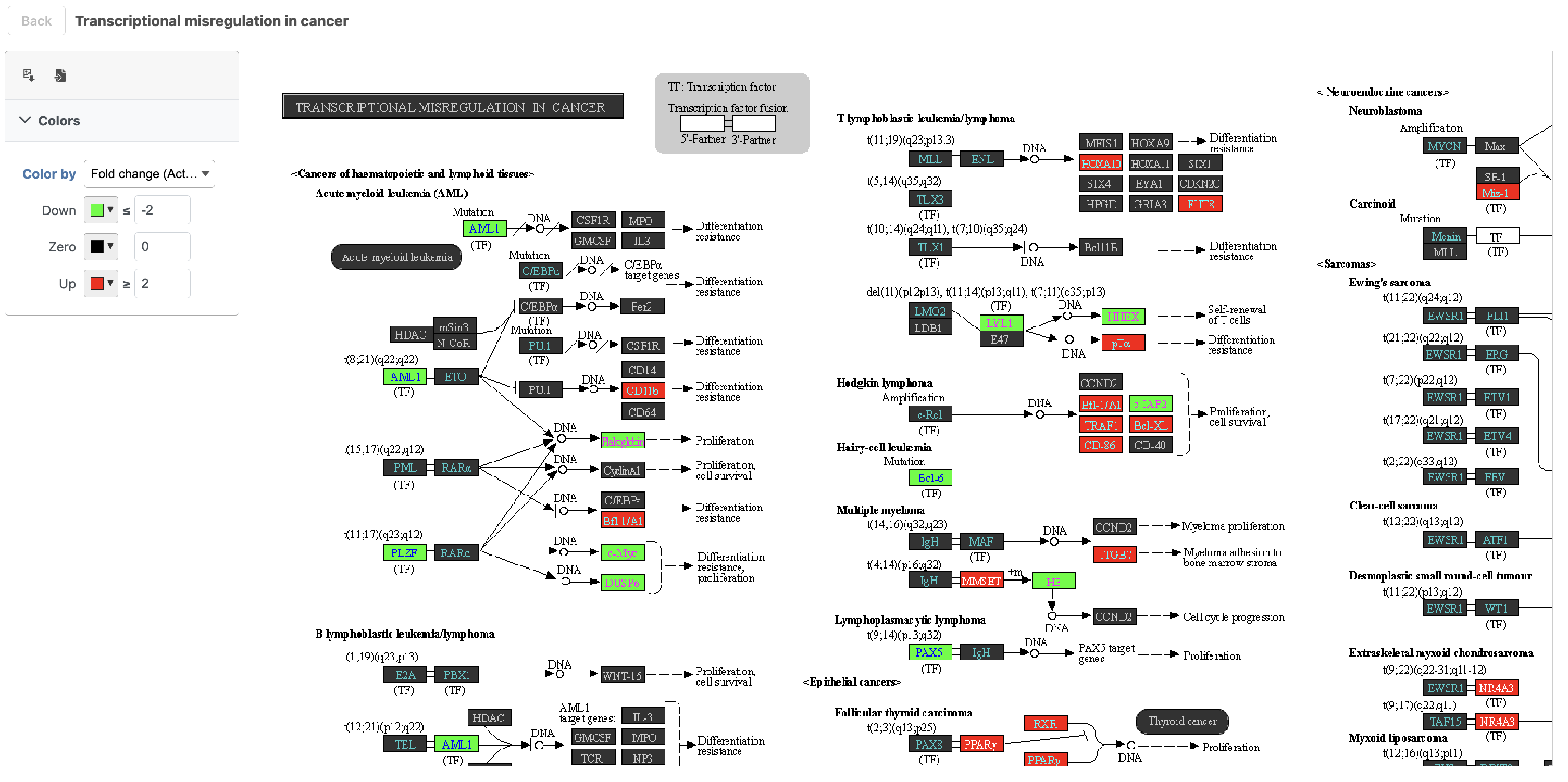 Image Added Image Added
|
| Numbered figure captions |
|---|
| SubtitleText | Final CITE-Seq pipeline |
|---|
| AnchorName | CITE-Seq final pipeline |
|---|
|
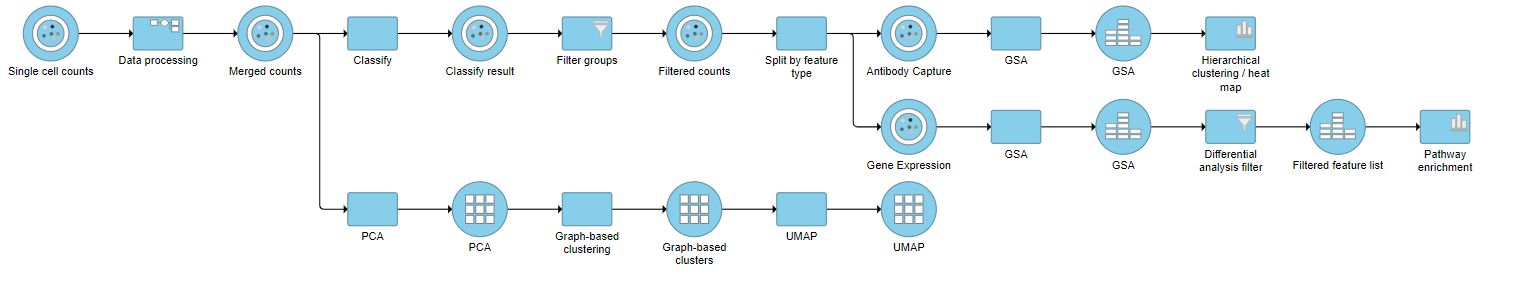 Image Removed Image Removed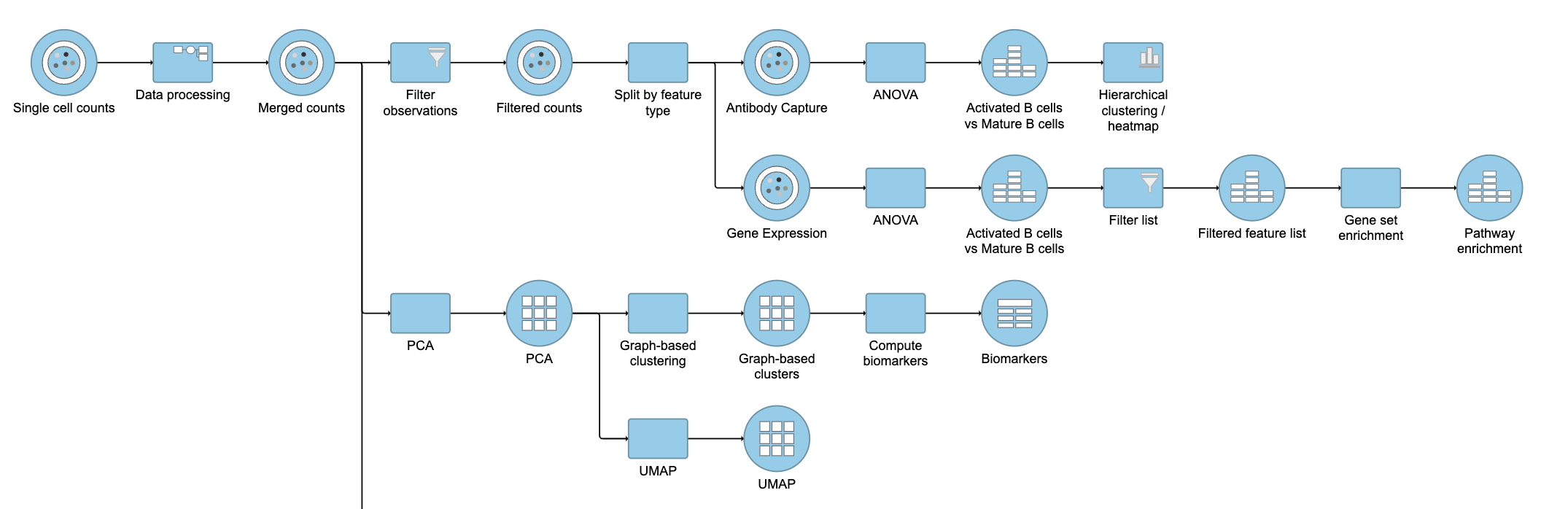 Image Added Image Added
|
...




































