Page History
...
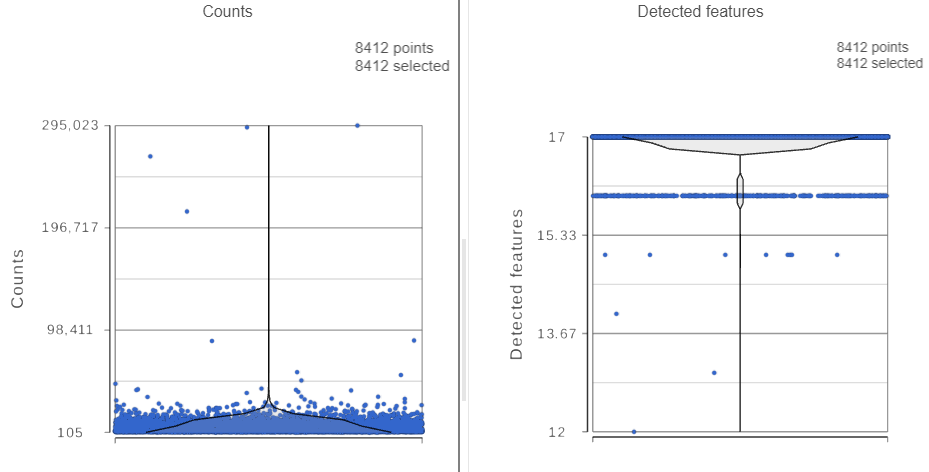
The Single cell QA/QC report opens in a new data viewer session. There are interactive violin plots showing the most commonly used quality metrics for each cell (Figure 3). For this data set, there are two relevant plots: the : the total count per cell and the number of detected features per cell. Each cell (Figure 3). Each point on the plots is a cell and the violins illustrate the distribution of values for the y-axis metric. Because mitochondrial transcripts are not present in protein data, this plot is not informative for this data set.
- Remove the % mitochondrial counts and the extra text box in the bottom right by clicking Remove plot in the top right corner of each plot (Figure 3)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
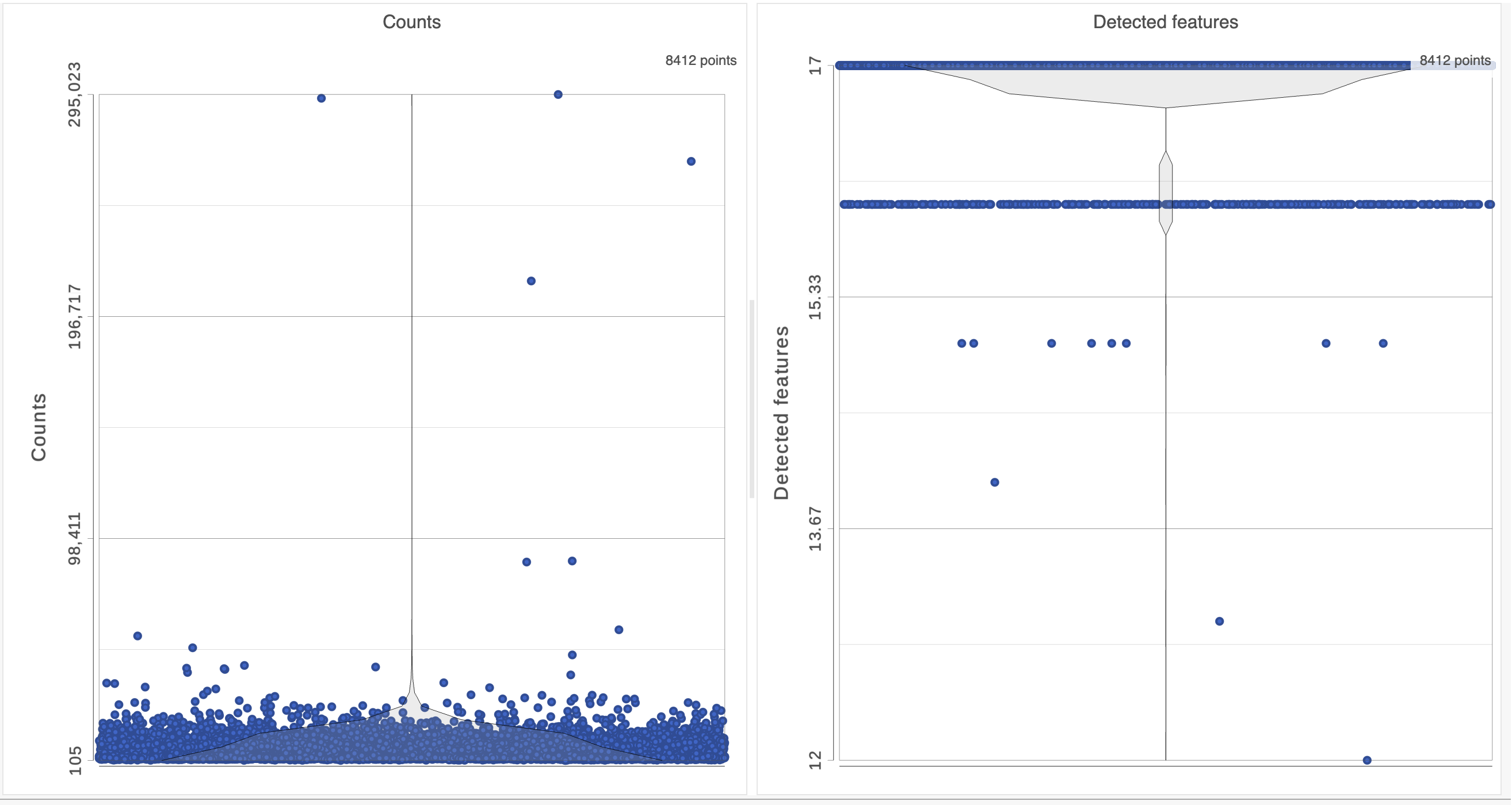
For this analysis, we will set a maximum counts threshold to exclude potential protein aggregates and, because we expect every cell to be bound by several antibodies, we will also set a minimum counts threshold.
- Select one of the plots on the canvas
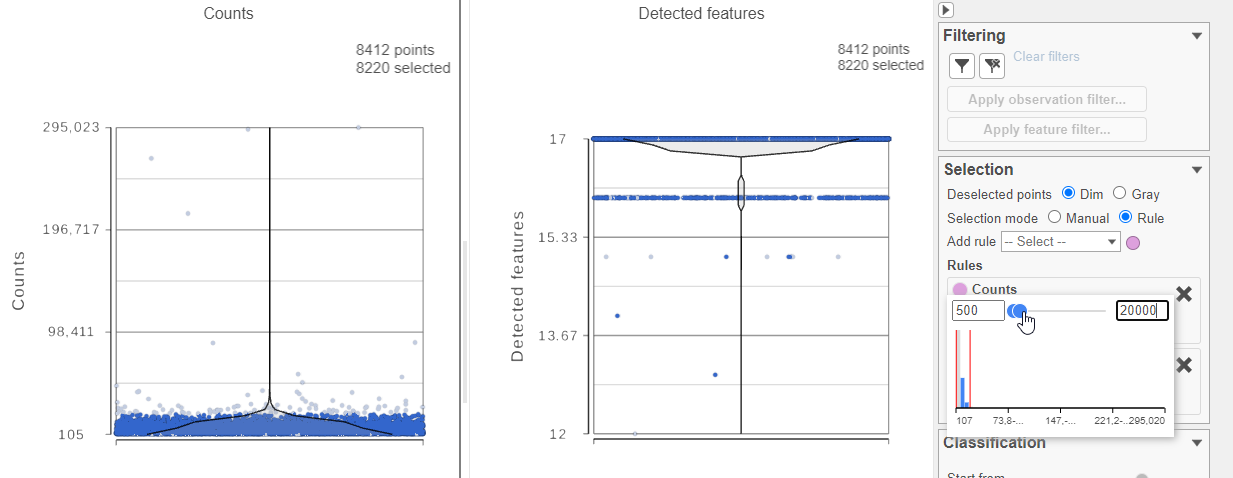
- In the Selection card Select & Filter icon on the rightleft under Tools, set the Counts threshold to keep cells between 500 and 20000 (Figure 4)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click in the Filteringunder Filter card on the right
- Click Apply observation filter...
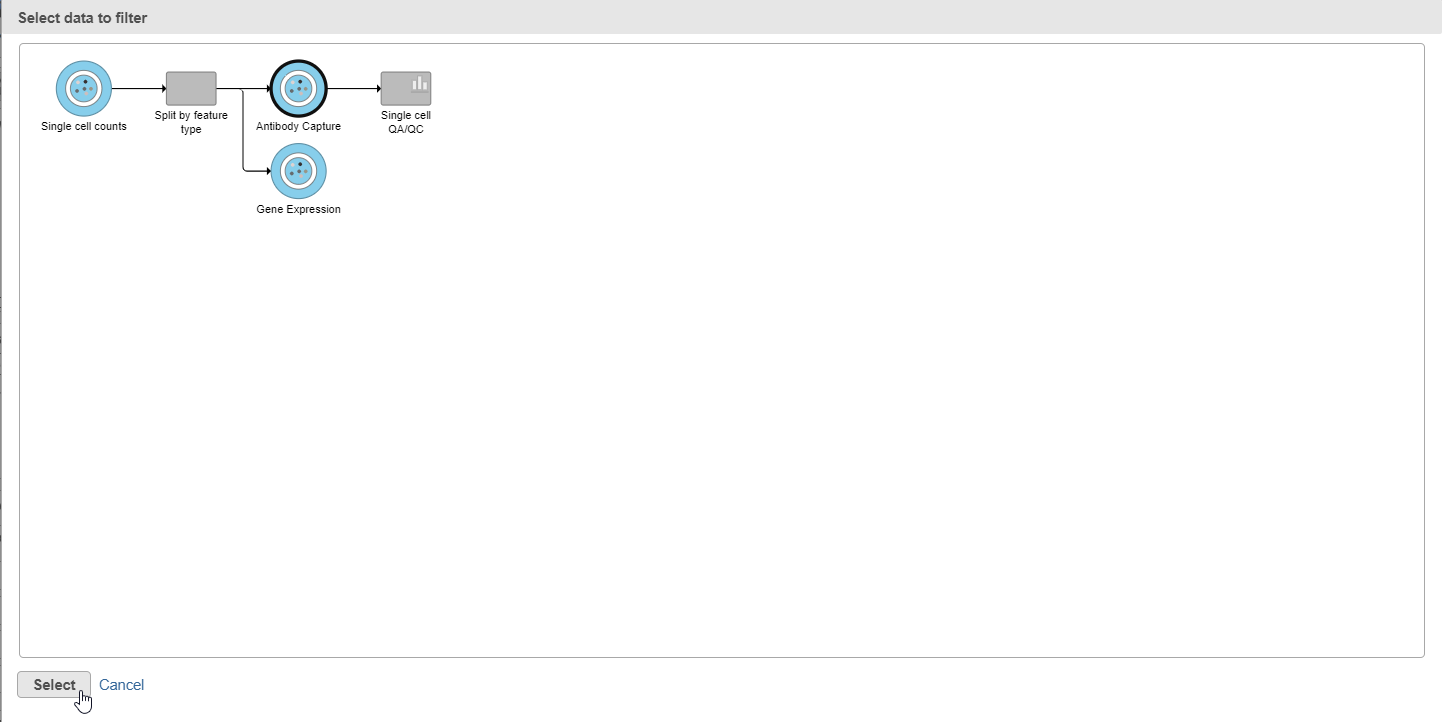
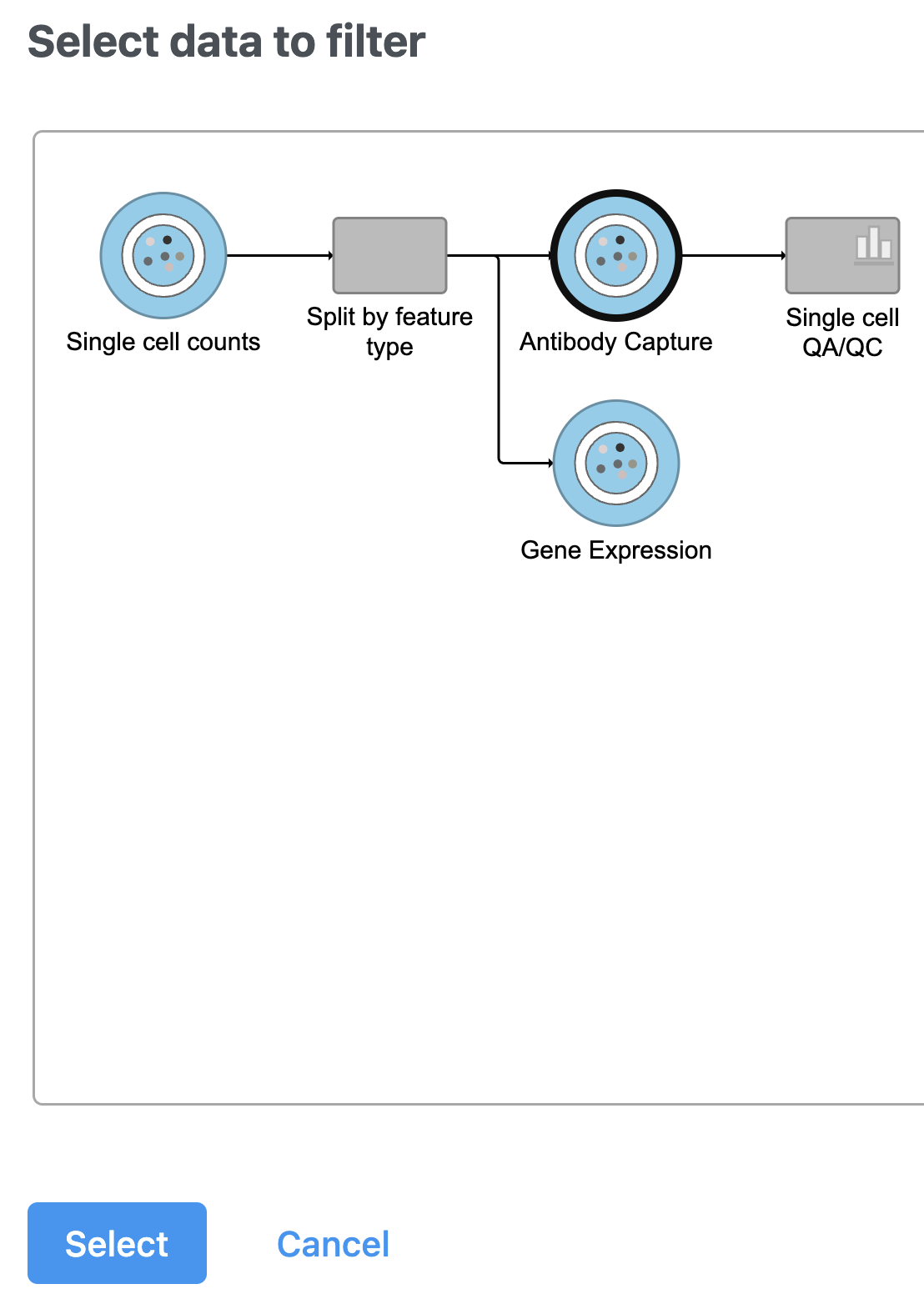
- Select the Antibody Capture data node as input in the pipeline preview (Figure 5)
- Click Select
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
You will see a message telling you a new task has been enqueued.
...
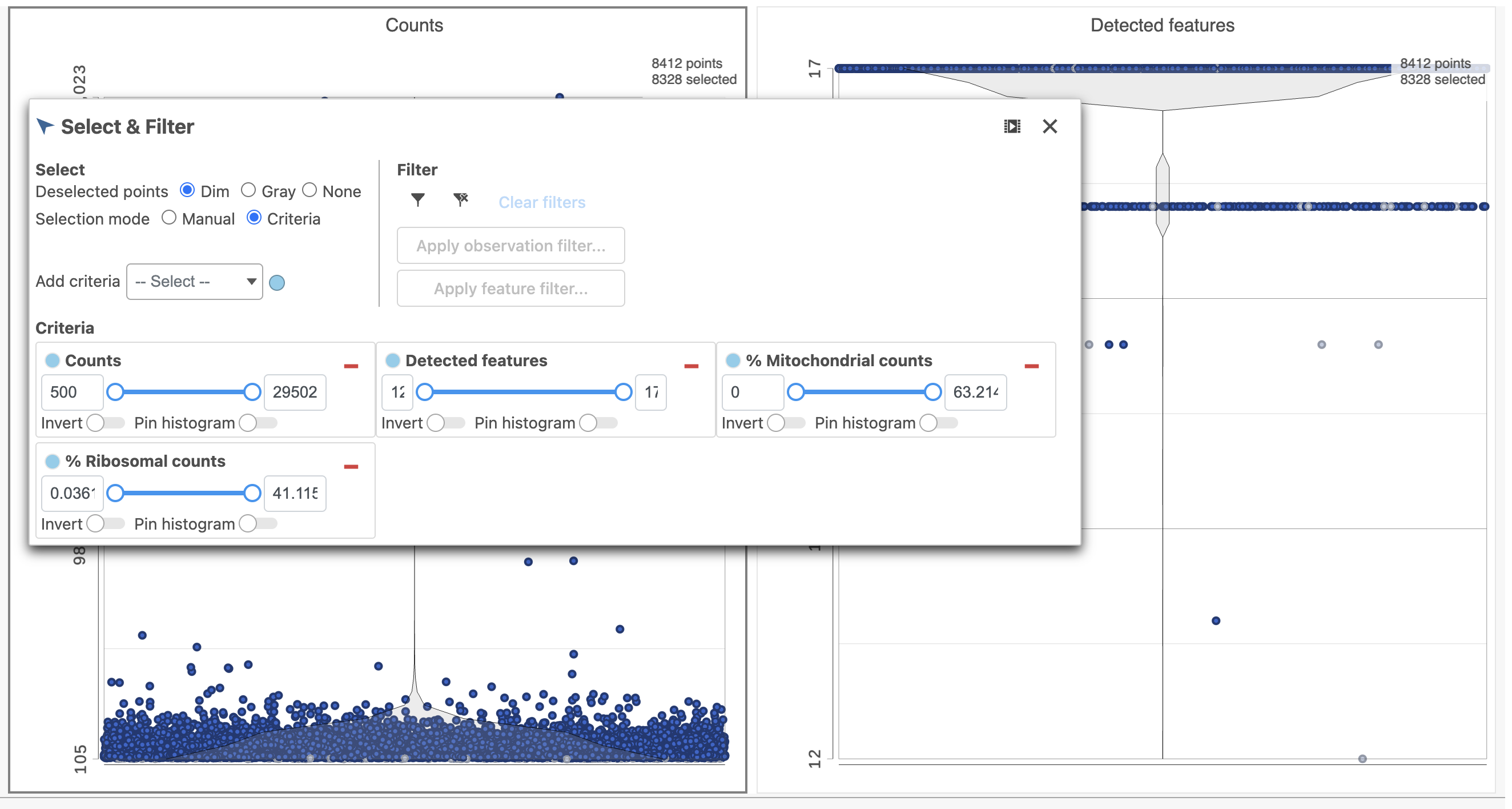
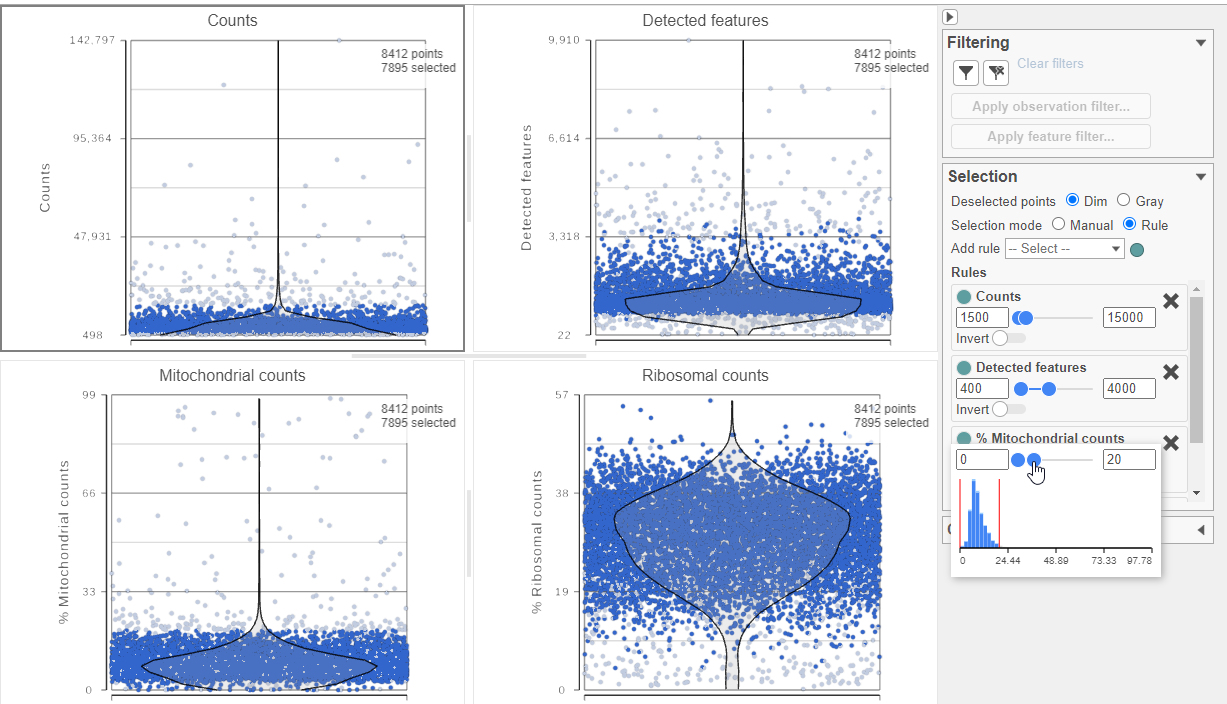
The task report lists the number of counts per cell, the number of detected features per cell, and the percentage of mitochondrial reads per cell in three , and the percentage of ribosomal counts per cell in four violin plots (Figure 6). For this analysis, we will set maximum and minimum thresholds for total counts and detected genes to exclude potential doublets and a maximum mitochondrial reads percentage filter to exclude potential dead or dying cells. There is no need to apply a filter based on the percentage of ribosomal counts in this tutorial.
- In the Selection card on the right, set the Counts threshold to keep cells between 1500 and 15000
- Set the Detected features to keep cells between 400 and 4000
- Set the % Mitochondrial counts to keep cells between 0% and 20% (Figure 6)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
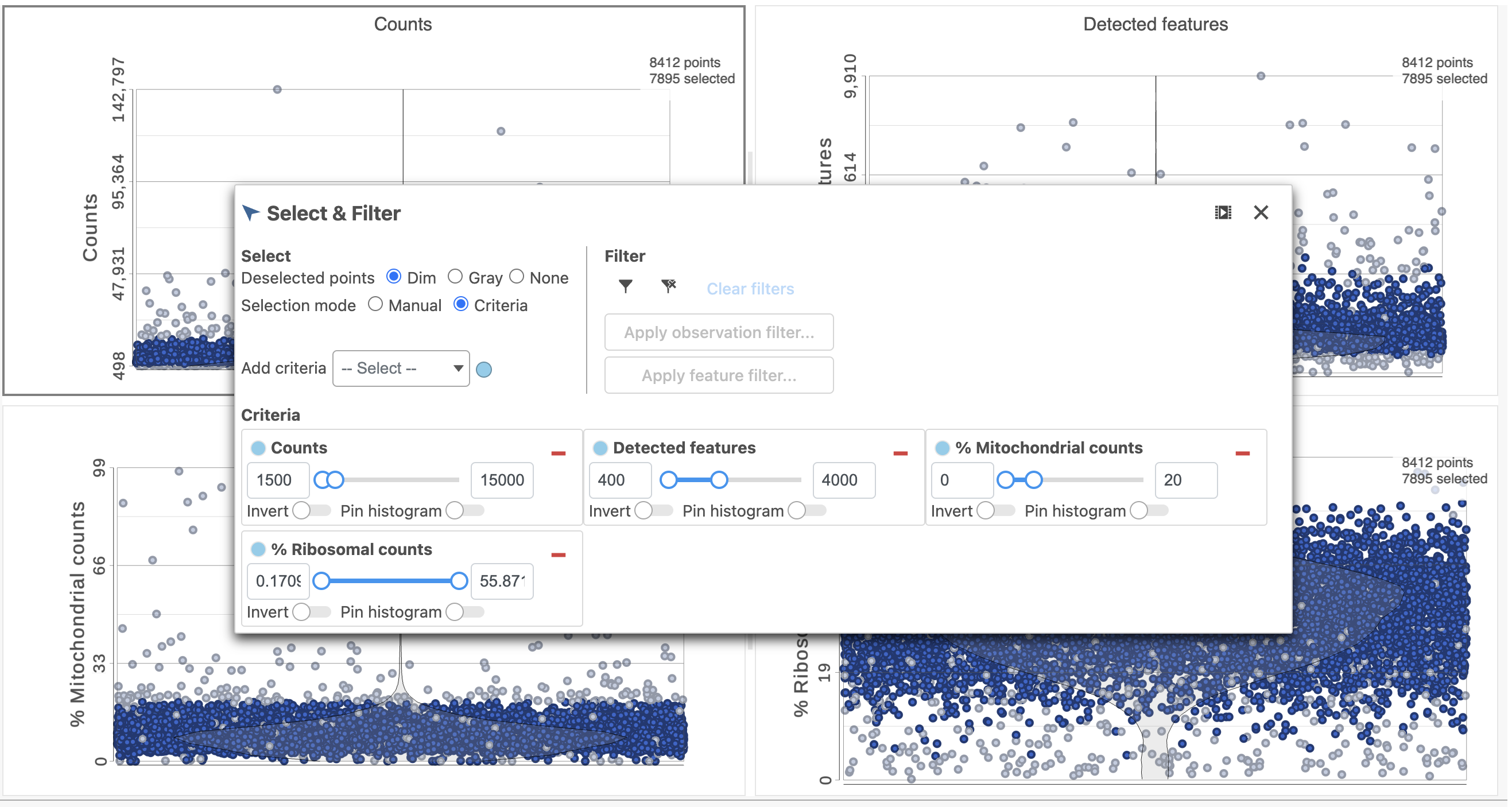
- Click in the Filtering card Filter on the right
- Click Apply observations filter
- Select the Gene Expression data node as input in the pipeline preview
- Click Select
- Click OK to dismiss the message about the task being enqueued
- Click the project name at the top to go back to the Analyses tab
- Your browser may warn you that any unsaved changes to the data viewer session will be lost. Ignore this message and proceed to the Analyses tab
...
- Click the Filtered counts data node produced by filtering the Antibody Capture data node
- Click Normalization and scaling in the toolbox
- Click Normalization
- Click the green button
- Click Finish to run (Figure 8)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
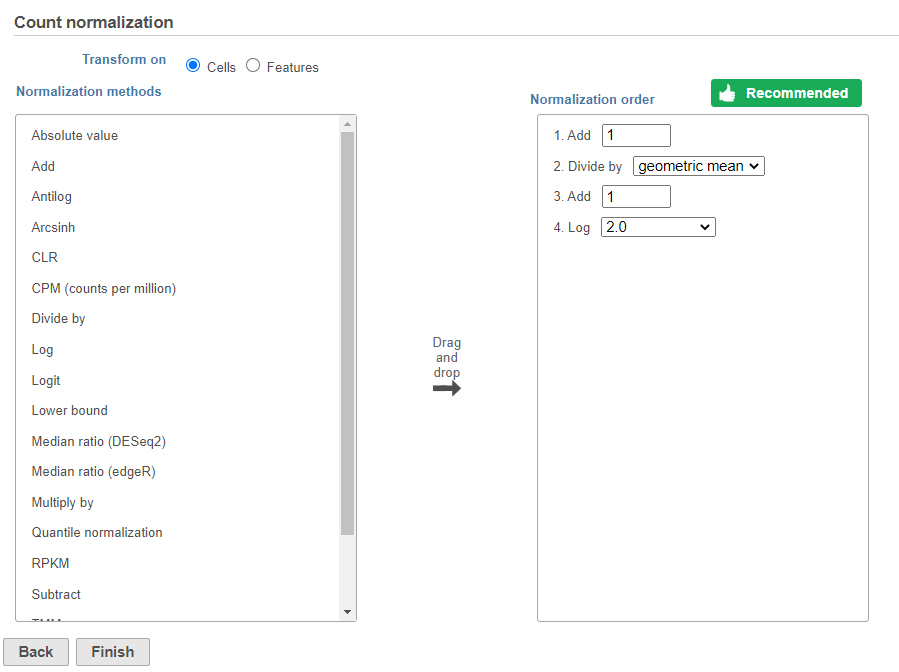
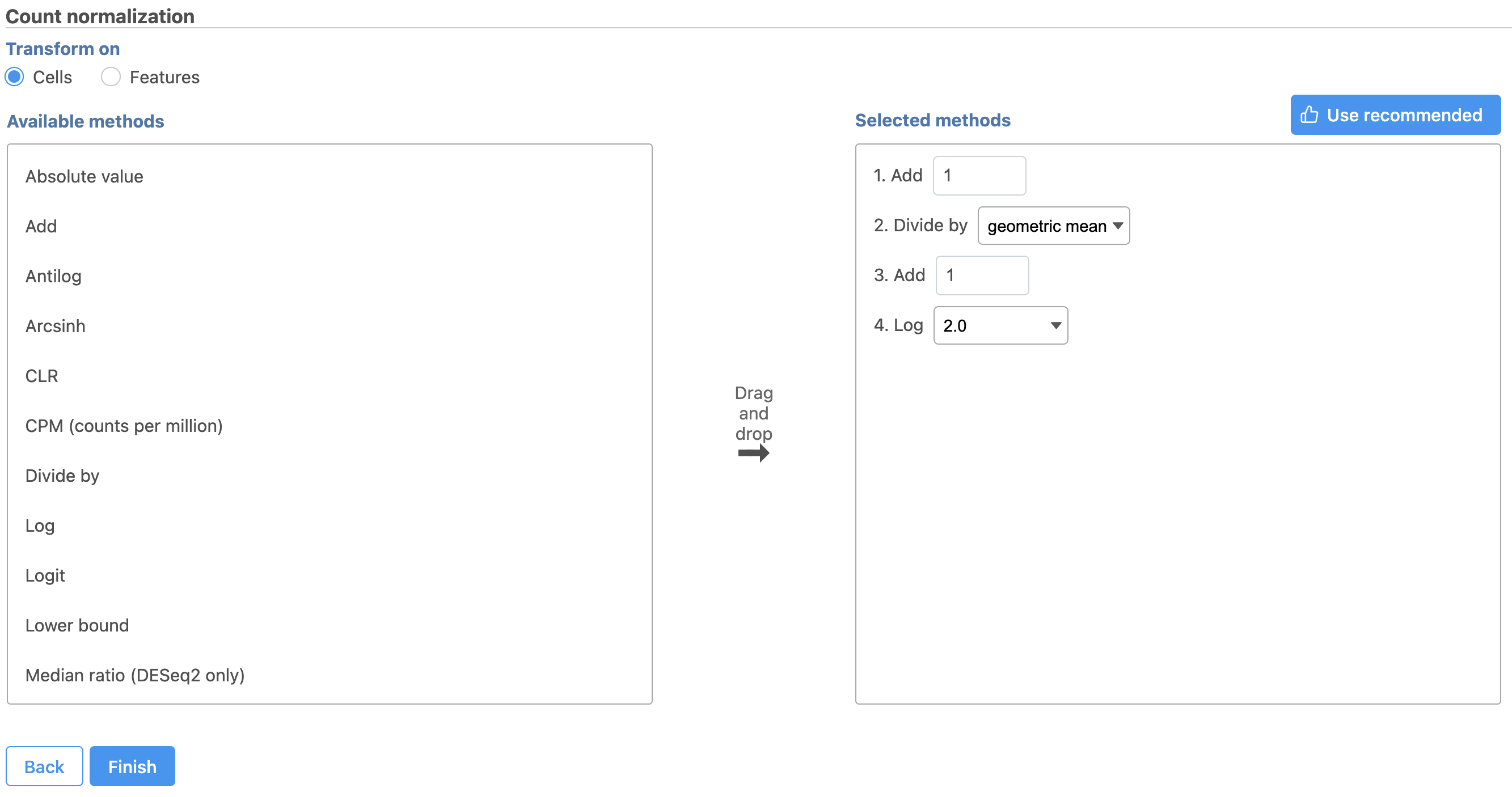
The recommended normalization for protein data includes the following steps: Add 1, Divide by Geometric mean, Add 1, Log base 2. This is a variant of Centered log-ratio (CLR), which was used to normalize antibody capture protein counts data in the paper that introduced CITE-Seq [1] and in subsequent publications on similar assays [2. 3]. CLR normalization includes the following steps: Add 1, Divide by Geometric mean, Add 1, log base e. Normalizing the protein data to base 2 instead of e allows for better integration with gene expression data further downstream. If you would prefer to use CLR, click and drag CLR from the panel on the left to the right. If you do choose to use CLR, we recommend making sure the gene expression data is normalized to the base e, to allow for smoother integration further downstream.
...
- Click the Filtered counts data node produced by filtering the Gene Expression data node
- Click the Normalization and scaling section in the toolbox
- Click Normalization
- Click the button
- Click Finish to run (Figure 9)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
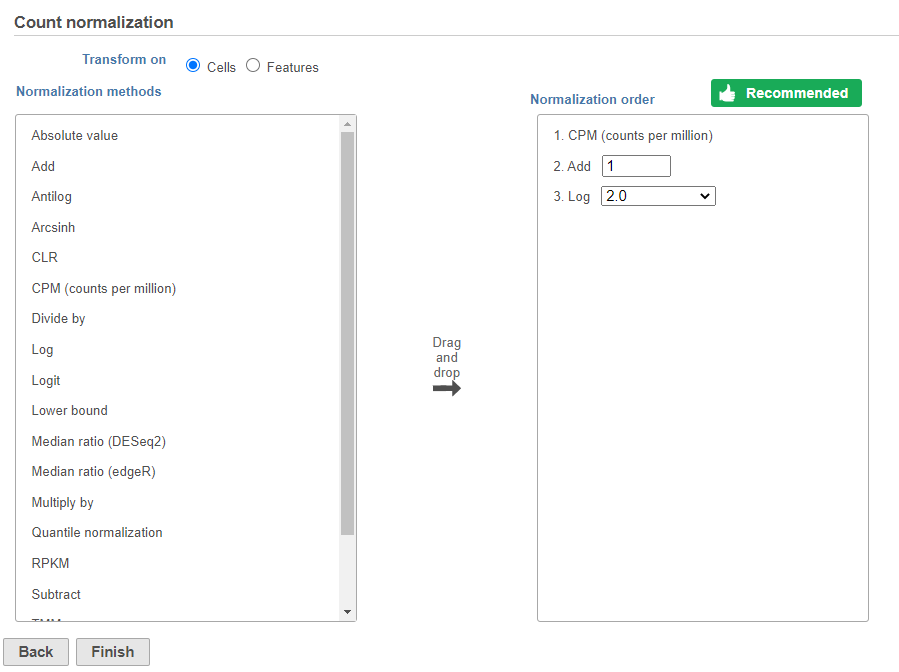
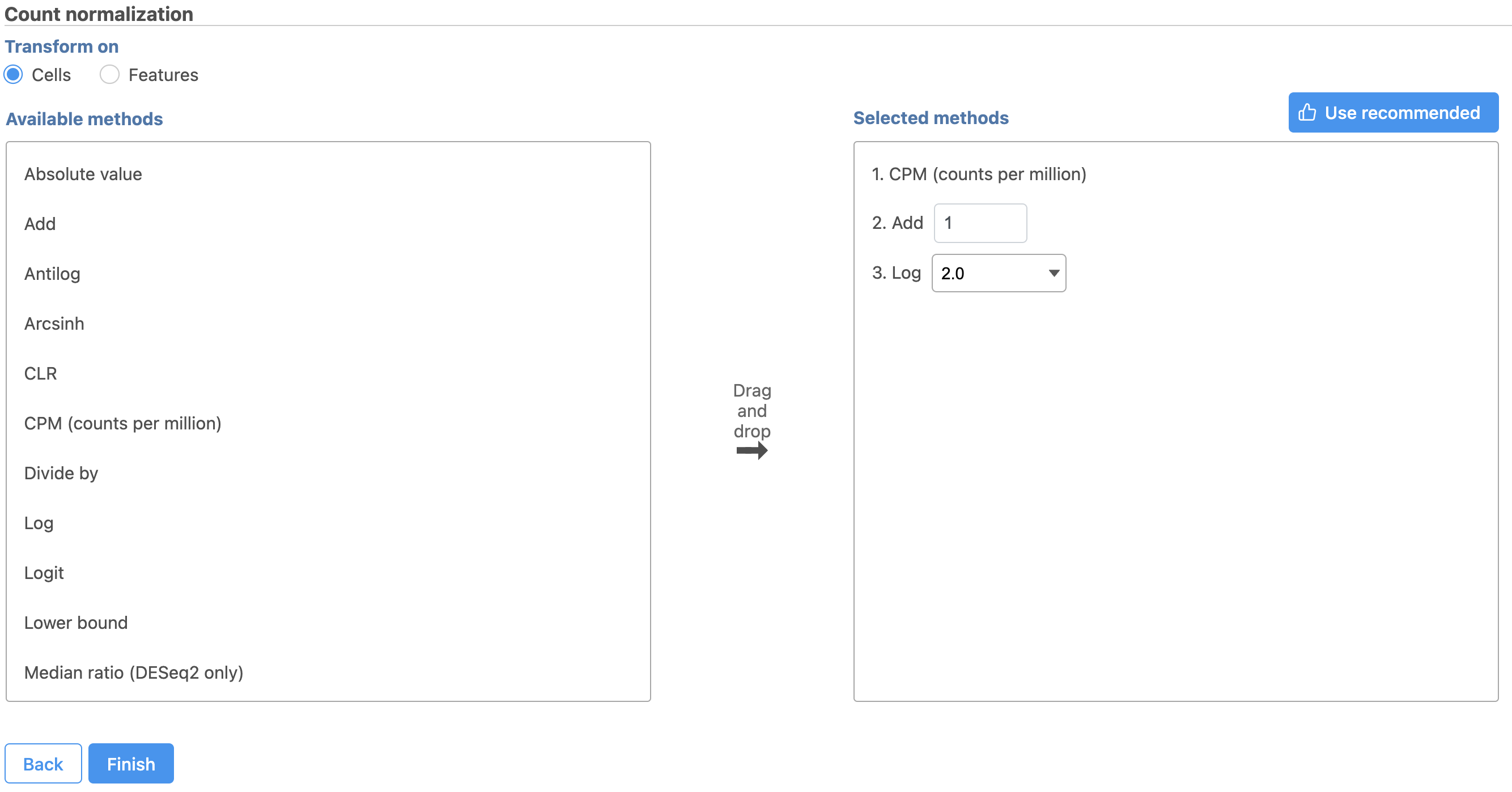
Normalization produces a Normalized counts data node on the Gene Expression branch of the pipeline (Figure 10).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
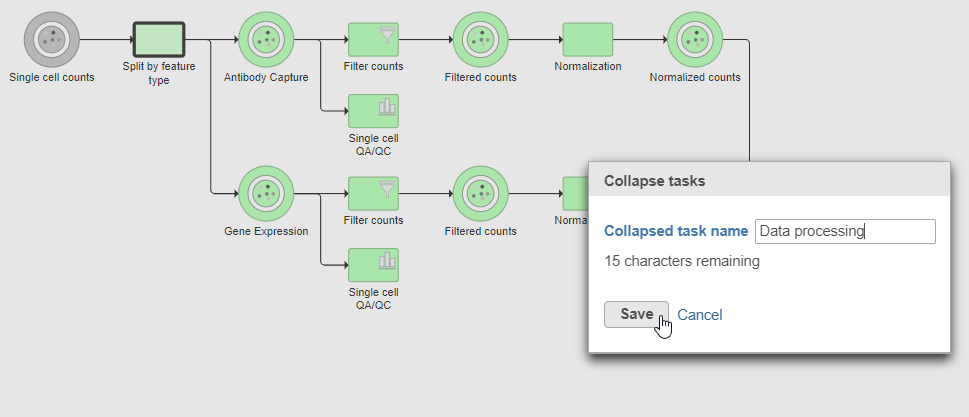
Tasks that can be selected for the beginning and end of the collapsed section of the pipeline are highlighted in purple (Figure 14). We have chosen the Split matrix task as the start and we can choose Merge matrices as the end of the collapsed section.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
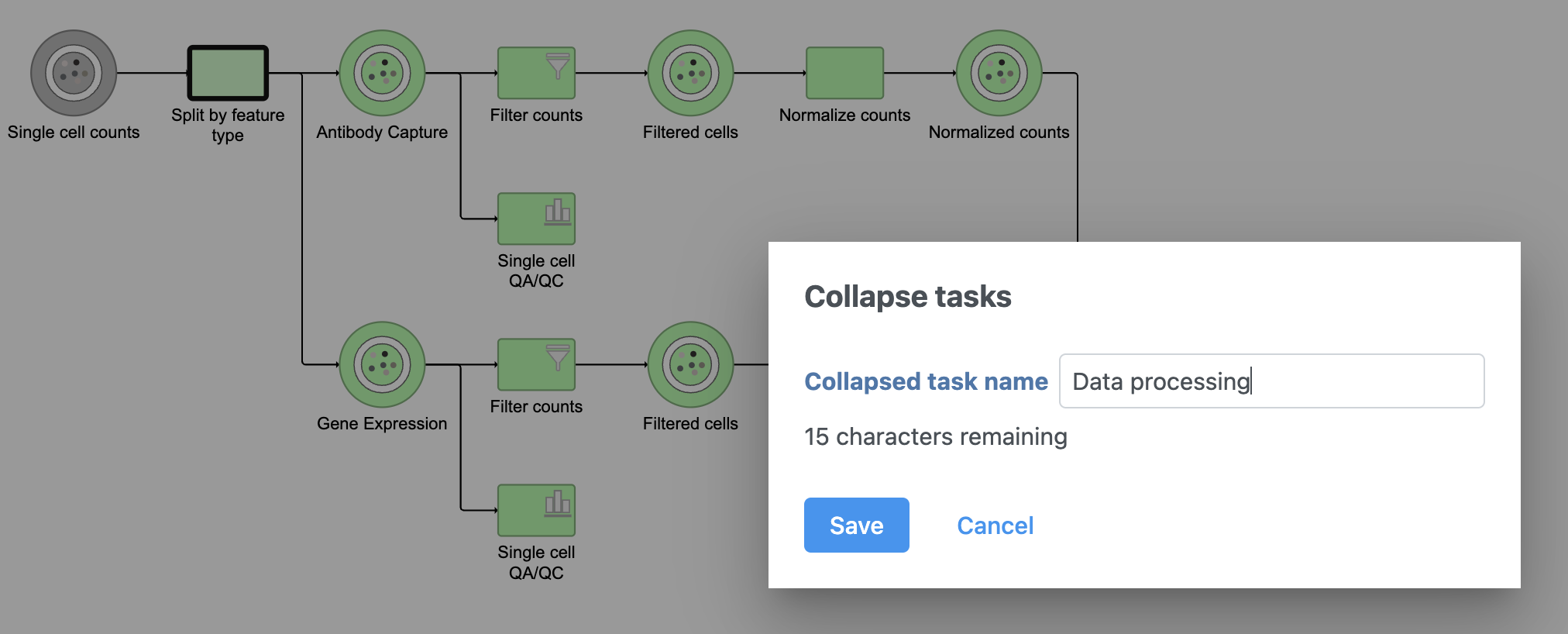
The new collapsed task, Data processing, appears as a single rectangular task node (Figure 16).
...