Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Create a new
...
Project
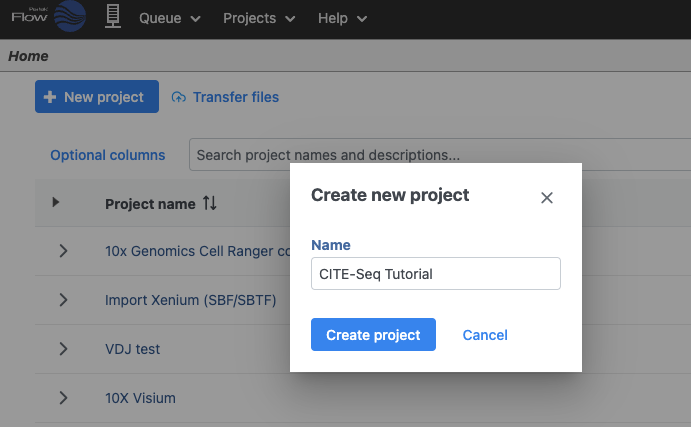
Let's start by creating a new project.
- On the Home page, click New project (Figure 1)
- Give the project a name
- Click Create project
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Import data
...
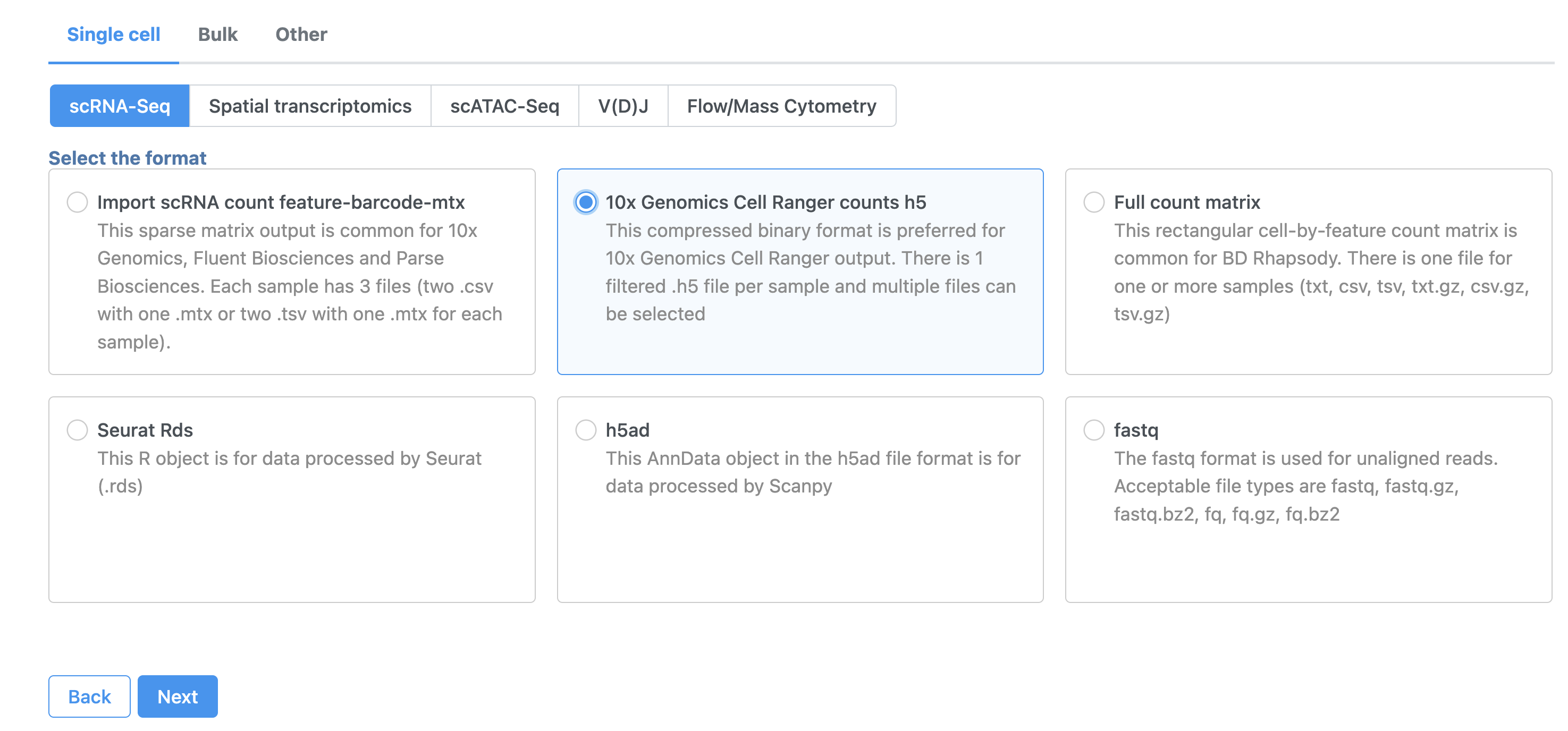
- In the Analyses tab, click Add data
- Click 10x Genomics Cell Ranger counts h5 (Figure 2)
- Choose the filtered HDF5 file for the MALT sample produced by Cell Ranger
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
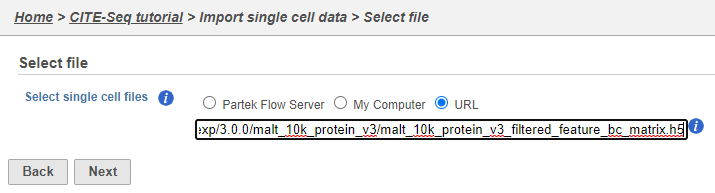
Move the .h5 file to where Partek Flow is installed , choose the Partek Flow Server radio button and using , then browse to its location. If you have downloaded the .h5 file to your local machine, choose the My computer radio button and upload the file through your web browser. Otherwise, select the URL radio button and paste in the following link (Figure ?):http://cf.10xgenomics.com/samples/cell-exp/3.0.0/malt_10k_protein_v3/malt_10k_protein_v3_filtered_feature_bc_matrix.h5
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
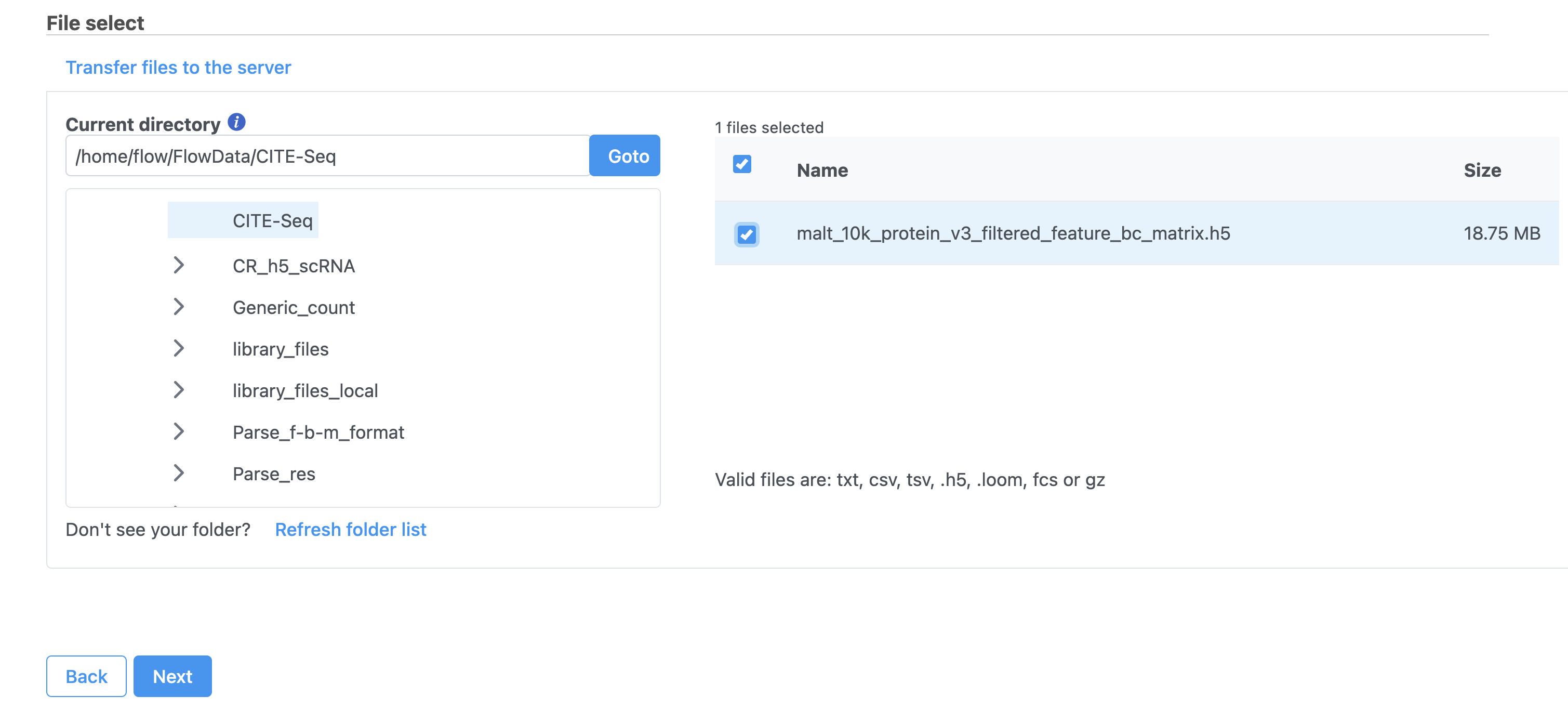
Note that Partek Flow also supports the feature-barcode matrix output (barcodes.tsv, features.tsv, matrix.mtx) from Cell Ranger. The import steps for a feature-barcode matrix are identical to this tutorial.
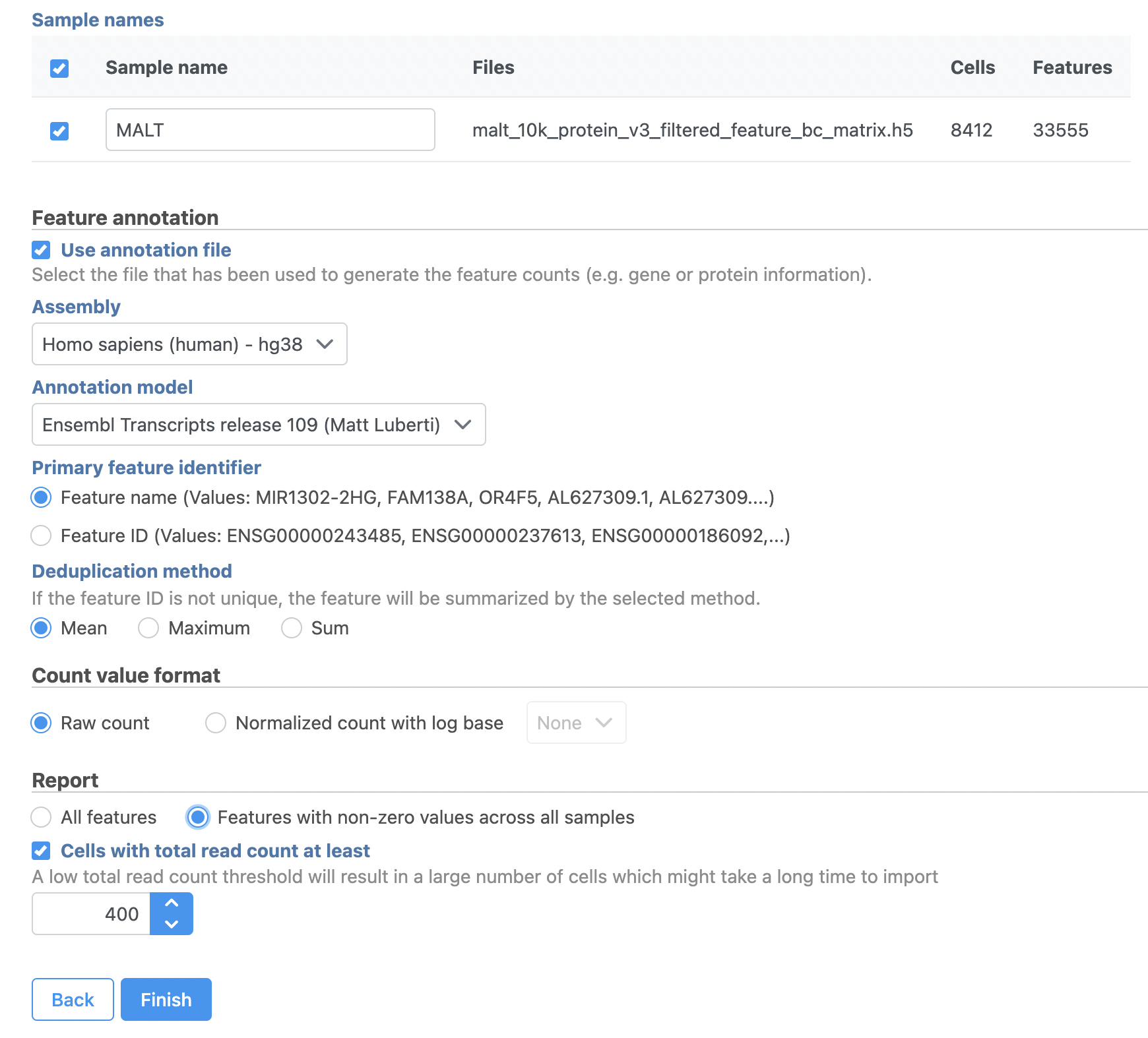
- Click Next
- Name the sample MALT (the default is the file name)
- Specify the annotation used for the gene expression data (here, we choose Homo sapiens (human) - hg38 and Ensembl Transcripts release
...
- 109). If Ensembl
...
- 109 is not available from the drop-down list, choose Add annotation and download it.
...
- Check Features with non-zero values across all samples in the Report section
- Click Finish (Figure
...
- 3)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A Single cell counts data node will be created under the Analyses tab after the file has been imported. We can move on to processing the data.
| Page Turner | ||
|---|---|---|
|
...