Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
The Trim tags task allows you to process unaligned read data with adaptors, barcodes, and UMIs using a Prep kit file that specifies the cofiguration of the adapters, barcodes, UMIs, and insert sequences these elements in your NGS reads.
Running Trim tags
- Click an Unaligned reads data node
- Click the Pre-alignment QA/QC section of the toolbox
- Click Trim tags
There are two parameters to configure - Prep kit and Keep untrimmed. Selecting Keep untrimmed will generate a separate unaligned reads data node with any reads that do not match the structure specified by the prep kit.
...
The output of Trim tags is a Trimmed reads data node. An additional Untrimmed reads data node will be generated if the Keep untrimmed option was selected.
...
Including a UMI in your prep kit will allow you to access a downstream task that uses UMI information , Deduplicate UMIsfor removing PCR duplicates. For more information about the Deduplicate UMIs task, please see our UMI Deduplication in Partek Flow white paper. While Note that while the UMI sequence will be trimmed, a record of the UMI sequence for each read is retained for use by this downstream taskstask.
When adding a UMI segment to your prep kit, you can specify the length of your UMIs (Figure 8).
...
Adding a barcode segment to a prep kit allows you to access downstream tasks that use barcode information, including Filter including Filter barcodes and Quantify barcodes to annotation model (Partek E/M). While the barcode sequence will be trimmed, a record of the barcode sequence for each read is retained for use by downstream tasks.
...
To set the barcode to an arbitrary segment of fixed length, choose Arbitrary and specify the barcode length (Figure 9).
...
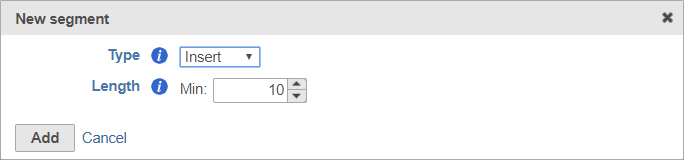
The insert is the sequence retained after trimming in the Trimmed reads data node. For example, in RNA-Seq, this would be the mRNA sequence. Every prep kit must include an insert segment. You can specify the minimum size of the insert section using the Length field (Figure 10). Reads shorter than the minimum length will be discarded.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Ordering segments
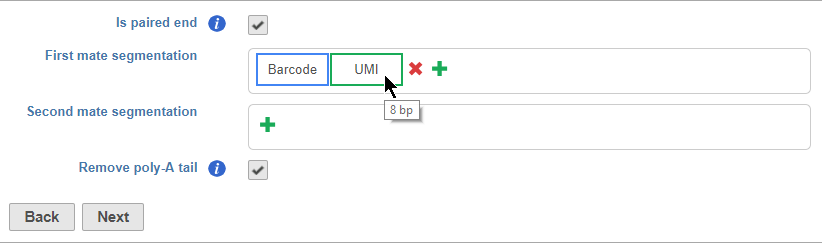
Segments are placed from 35' to 53' in the read in the order they are added. You should add the 35' segment first and add additional elements in order of their position in the read. Segments will appear in the Segmentation sections as they are added. You can mouse over a segment to view its details (Figure 11).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Custom prep kit example
For example, the expected read structure (Figure 12) and a completed prep kit for a standard Drop-seq library prep are shown below (Figure 13).
...