Page History
...
To run Hashtag demultiplexing, your data must meet the following criteria:
- Cell Hashing features must have a different data type than your gene expression or protein expression data
- Cell Hashing data should be normalized with CLRData node contains number of features less than number of observations
- Data node must be output from normalization task (recommended normalization method for hashtag is CLR)
If you are processing your FASTQ files in Partek Flow, be sure to specify a different Data type for your Cell Hashing FASTQ files on import than the FASTQ files for your gene expression and any other antibody data.
...
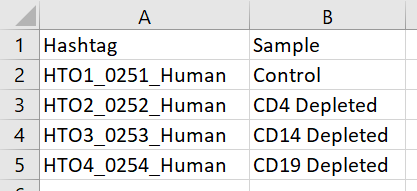
If you want to specify sample IDs instead of using hashtag feature IDs as the sample IDs, you will need to prepare a tab-delimited text file (.txt) with hashtag feature IDs in the first column (Hashtag) and the corresponding sample IDs in the second column (Sample) (Figure 1). A header row is required.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Overview
Content Tools