Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Motif detection tasks allow to extract identify transcription factor binding motifs from in peak region regions generated from by ChIP-seq Seq and ATAC-Seq data. It allows Partek Flow includes tasks that allow users to search for know known motifs from provided a database as well as to detect noval novel motifs. These tasks are available when click on region data nodecan be invoked on data nodes with genomic regions.
Search for know motifs
Given a set of genomic regions, sequence motif can be search based on a string provided by user or alignment matrix from a database like JASPAR. Click on a peak data node, choose Search for known motifs from Motif detection section on the pop-up menu (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
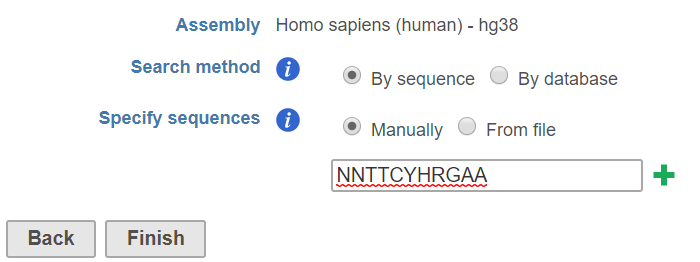
When choose the search method by sequence, there are two options:
...