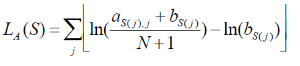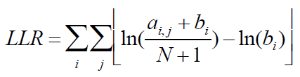Page History
...
Let M be a motif of length L consisting of N motif instances.Let A be a 4XL alignment matrix such that ai,j is the count of letter i at position j. Let Bi be the background frequency of letter i (calculated as the number of nucleotides i in the regions divided by the total oligonucleotides in the regions). Let S be a sequence of length L. The score of S given the alignment matrix is
Equation 1
Let h be the maximum of LA. The quality score of a sequence is calculated as QA(S) =LA(S)/h. A quality score of 1 corresponds to a sequence with the most likely base at each position of the alignment matrix. User will specify a threshold QA. all sequences that have a score TA>QA *h will be reported.
The report contains summary and detail tabs:
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
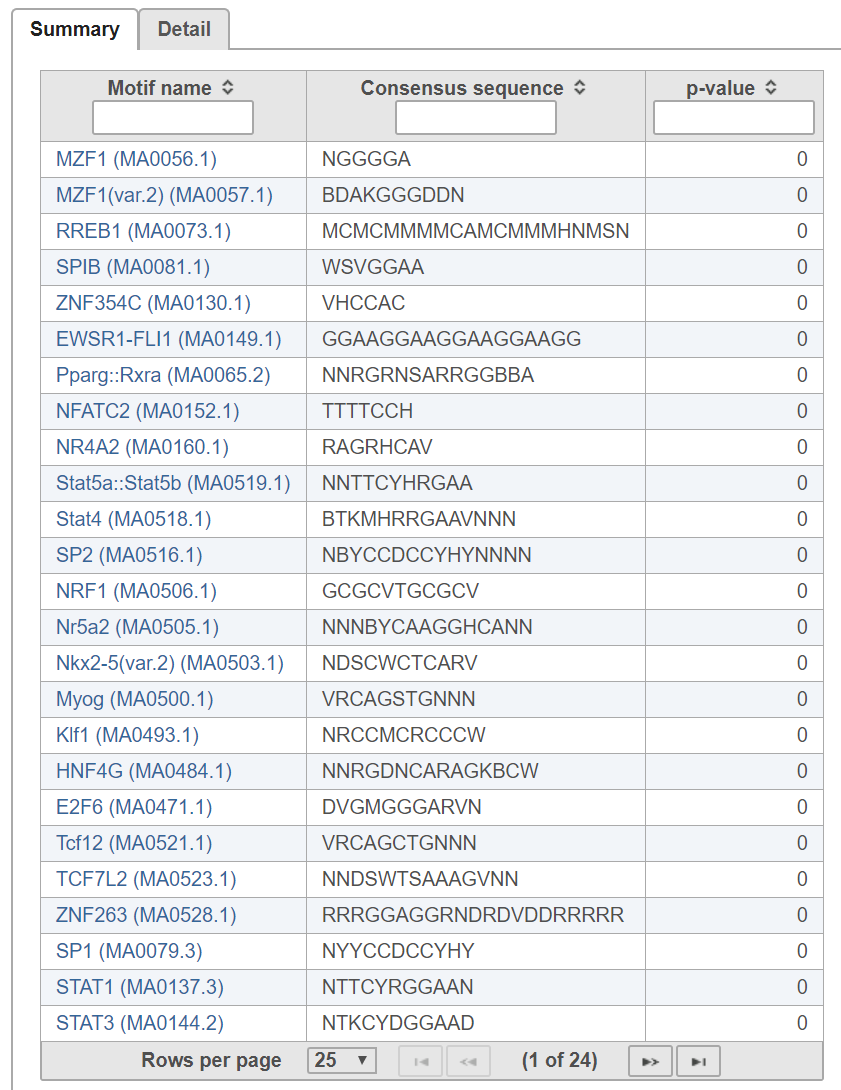
The probability PExpected of a sequence having a score above TA is calculated under the assumption that the base are i.i.d. according to the background distribution B. Let NTrials be the number of sequences compared to the alignment matrix. The expected number of occurrences of the motif in the regions is PExpected * NTrials. The p-value of observing NActual instances with a score above TA is calculated based on the binomial distribution, where NTrials is the number of trials and PExpected is the probability of success. A low p-value indicates that the regions are enriched with instances of the motif.
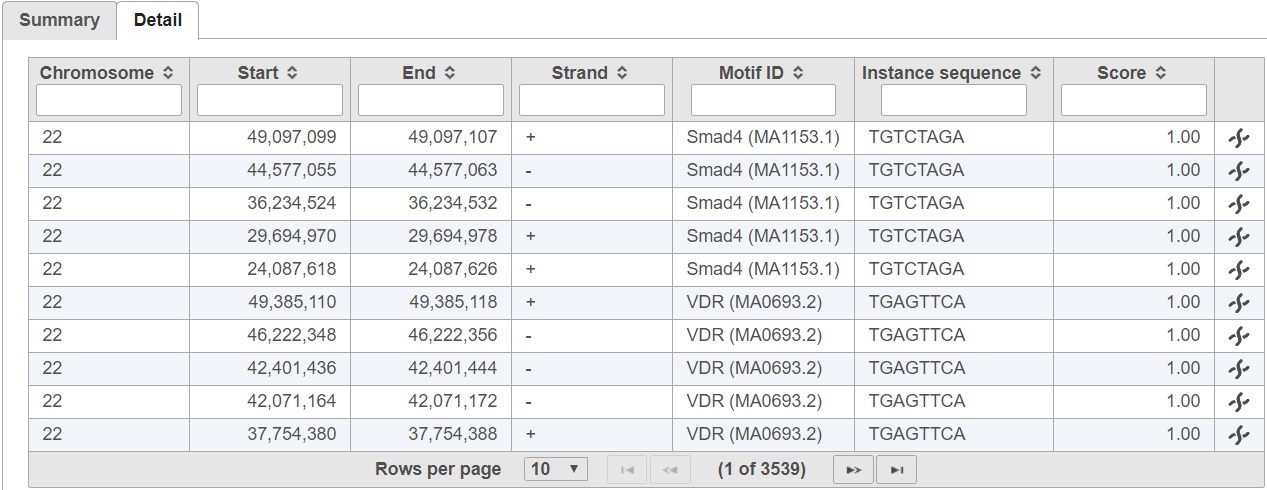
The In the detail table, each row is a location containing a motif sequence with quality score (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Detect de novo motifs
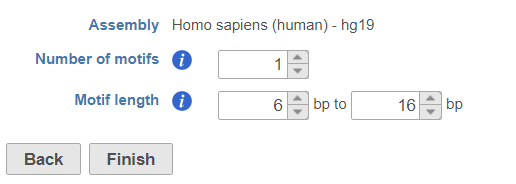
Click on Peak data node, select Detect de novo motifs from Motif detection section in the pop-up menu (Figure 14), specify the number of motifs to detect and the length of the motifs.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Equation 2
This is done by repeating the below two steps until convergences:
...
The Gibbs sampler is run on a range of the motif sizes specified by the user. The motif with the greatest average LLR (LLR /length) is returned. To find N motifs ina set of regions, the Gibbs sampling method is run N times. The motif instances found from the previous run of the Gibbs sampler are removed before performing the next run.
The report contains summary and detail tabs:
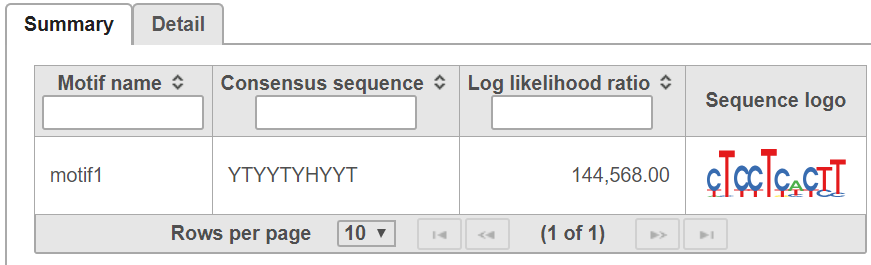
In summary table, it contains motif consensus sequence and sequence logo (Figure 5):
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The detail table contains the locations of motif sequence (Figure 6):
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Reference
- Hertz, GZ., & Stormo, G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 1999, 15, 563-577
- Schug, J., & Overton, C.G. TESS, Transcription Element Search Software on WWW. Technical Report CBIL-TR-1997-1001-v0.0, of the Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania, 1997
Rate Macro allowUsers false
...