Page History
...
The Number of principal components should be set based on the your examination of the Scree plot and component loadings table. The default value of 100 is likely exhaustive for most data sets, but may introduce noise that reduces the resolution number of clustering (fewer clusters identified)clusters that can be distinguished.
- Click Apply
- Click Finish to run
...
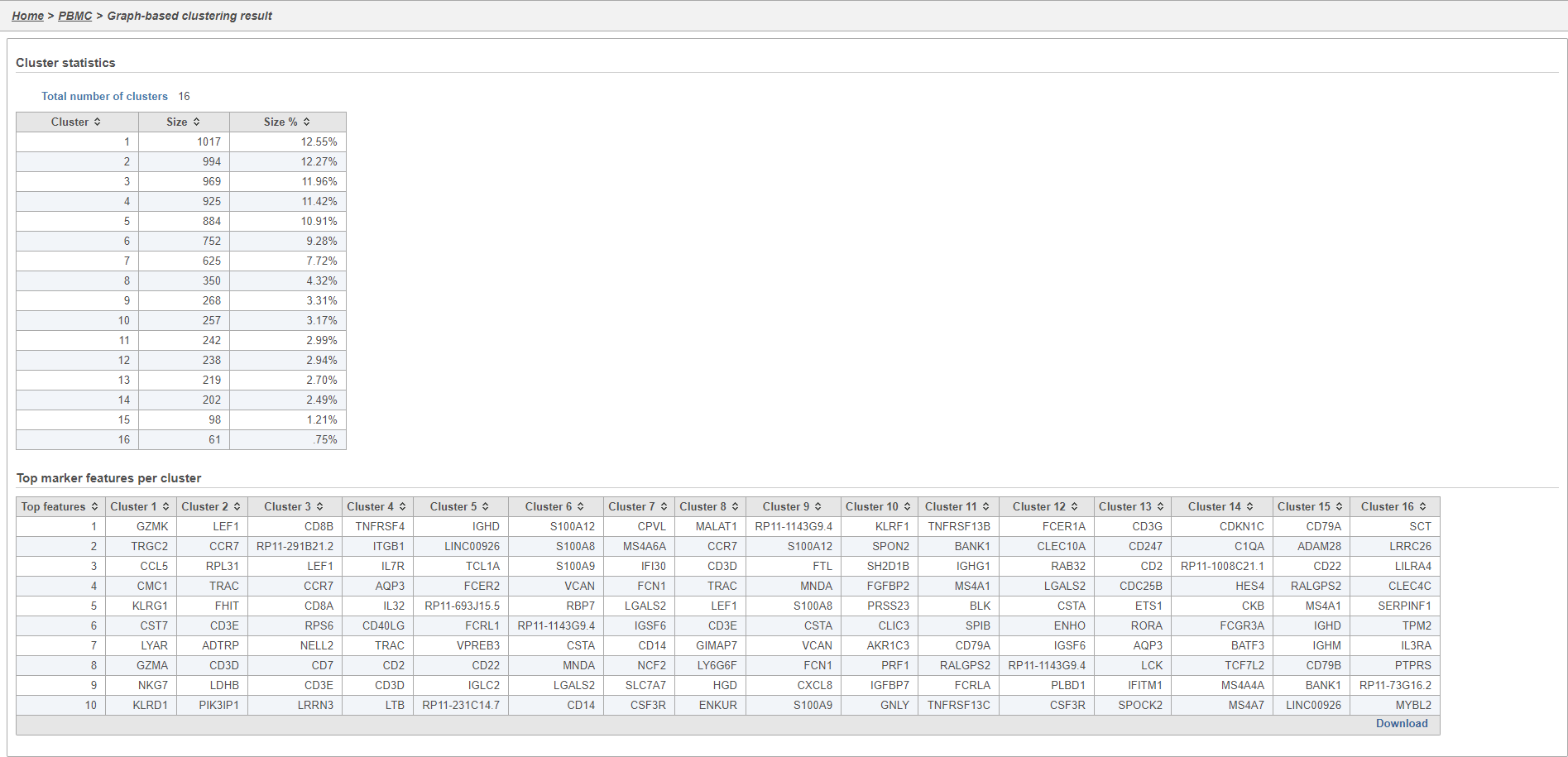
The Graph-based clustering task report lists the number of clusters and what proportion of cells fall into each cluster. It also includes a cluster biomarkers table. This lists the top-10 genes that distinguish each cluster from the others (Figure 9). These are calculated using an ANOVA test comparing the cells in each group to all the other cells, filtering to genes that are 1.5 fold upregulated, and sorting by ascending p-value. This ensures that the top-10 genes of each cluster are highly and disproportionately expressed in that cluster.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...