Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Differential expression analysis can be used to compare cell types. Here, we will compare malignant glioma and oligodendrocyte cells to identify genes differentially regulated in malignant glioma cells from the Oligodendroglioma subtype. This comparison is of interest because malignant glioma cells in Oligodendroglioma are thought to originate from oligodendrocytes.
...
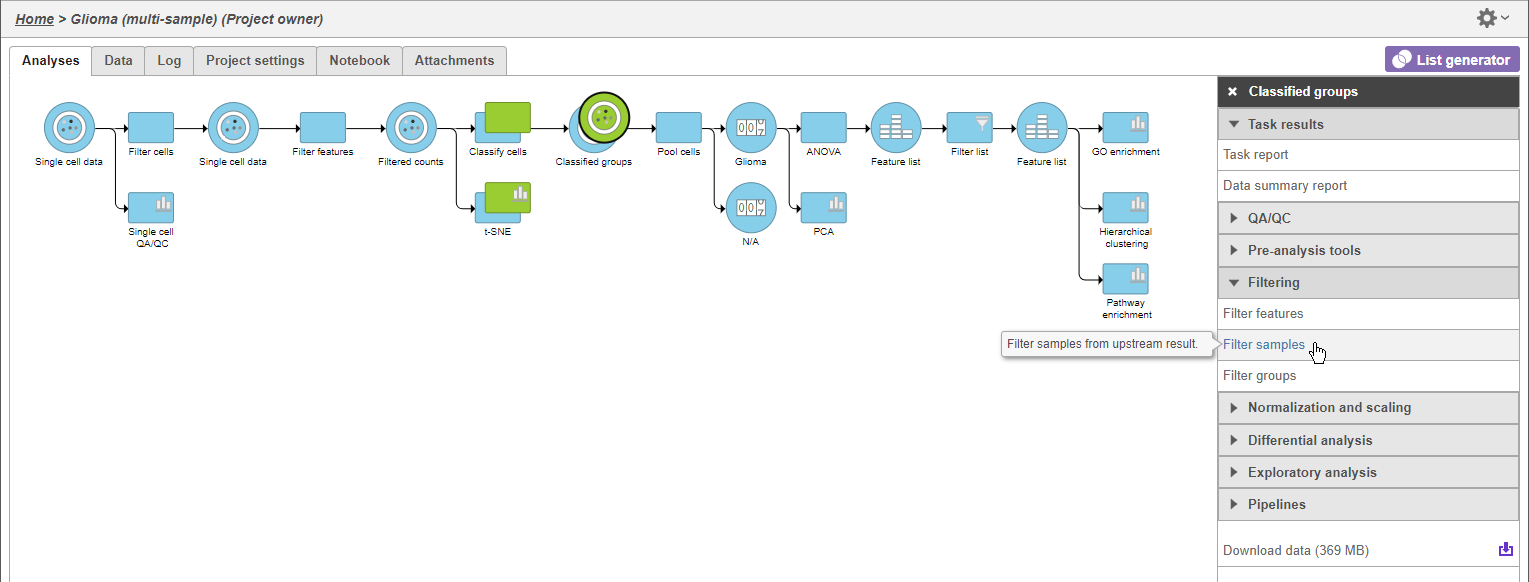
To analyze only the Oligodendroglioma subtype, we can filter the samples.
- Select Click the green Normalized counts Classified groups data node
- Select Filter samples from the Filtering section of Click Filtering in the task menu
- Click Filter samples (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
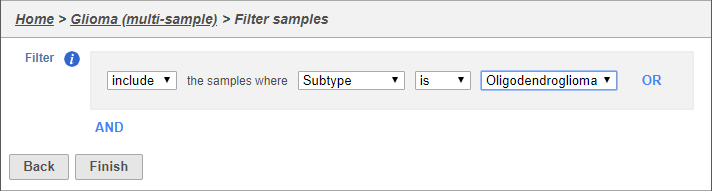
The filter lets us include or exclude samples based on sample ID and attribute (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Set the filter to Include samples where Type Subtype is Oligodendroglioma
- Select AND
- Set the second filter to Exclude samples where Classification is Microglia
- Select Click Finish to apply the filter
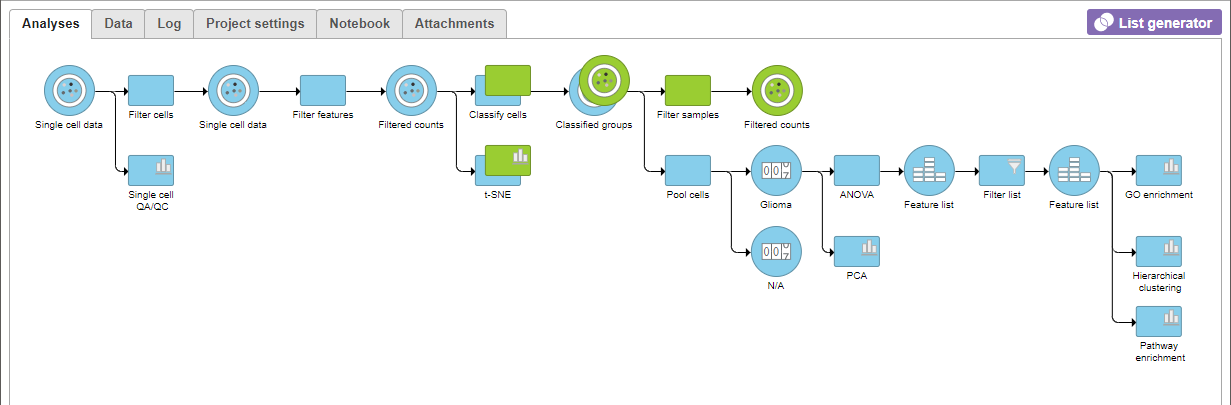
A Filtered Counts data node will be created with only cells that are from Oligodendroglioma samples (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Filtering groups
Because we are only interested in analyzing glioma and oligodendrocyte cells, we will filter out microglia cells using the groups filer.
- Click the green Filtered counts data node
- Click Filtering in the task menu
- Click Filter groups
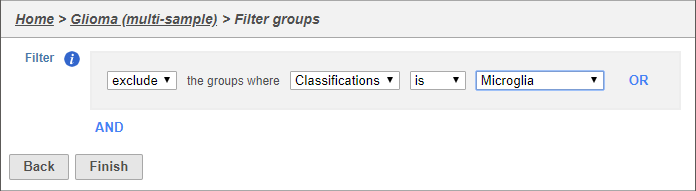
This filter lets us include or exclude cells based on classifications or other cell-level attributes.
- Set the filter to Exclude groups where Classification is Microglia (Figure 4)
- Click Finish to apply the filter
A Filtered Counts
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A Filtered Counts data node will be created with only glioma and oligodendrocyte cells that are from the Oligodendroglioma samples and are classified as either malignant or oligodendrocyte.. The Filtered groups task must complete before we can proceed to identifying differentially expressed genes.
Identify differentially expressed genes
- Select Click the second green Filtered counts data node
- Select Detect differential expression (GSA)
...
- Click Differential analysis in the task menu
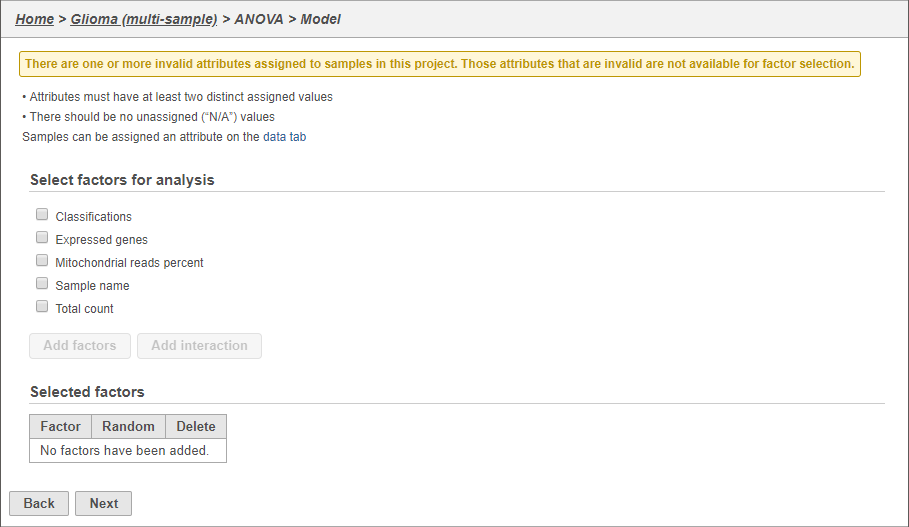
- Click ANOVA (Figure 5)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click Classification
- Click Add factors
- Click Next
Next, we will set up a comparison between malignant cells gliioma and oligodendrocytes in Oligodendroglioma.
- Select Malignant and Oligodendroglioma in the top panels
- Select Oligodendrocytes in the bottom panels
Because we are analyzing sparse data, we need to relax the default low expression filter. By default the low expression filter is set a minimum of 1 average normalized read; here, we will turn it off.
...
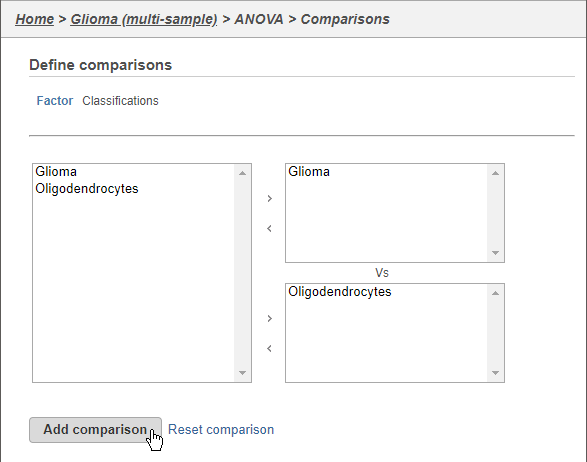
- Click Glioma
- Click the arrow to move it to the top-right panel
- Click Oligodendrocytes
- Click the arrow to move it to the top-right bottom-right panel
- Click Add comparison (Figure 8)
This will set up fold calculations with glioma as the numerator and oligodendrocytes as the denominator.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click None in the Read count normalization section
- Click Finish to run the ANOVA
A green Feature list node will be generated containing the results of the ANOVA.
- Double-click the green Feature list node to open the ANOVA report
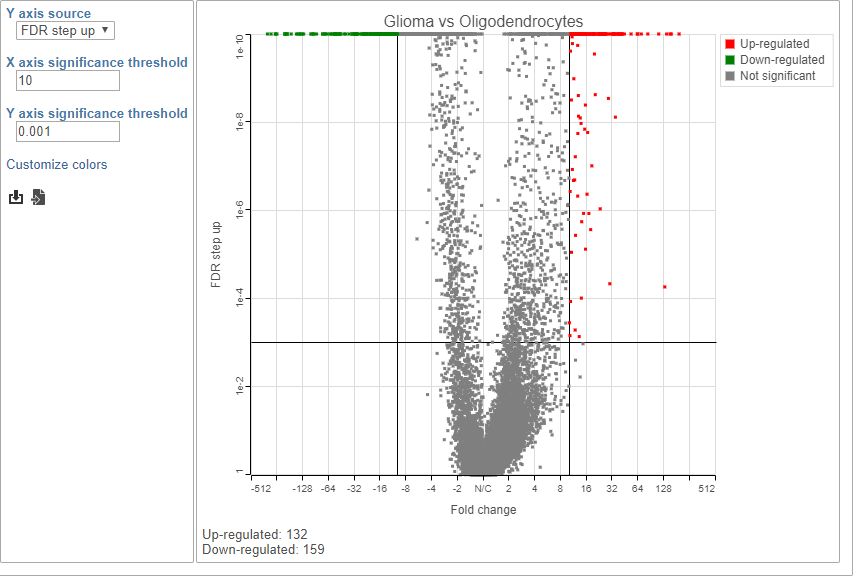
Because of the large number of cells and large differences between cell types, the p-values and FDR step up values are very low for highly significant genes.
- Click to view the Volcano plot
- Choose FDR step up from the Y axis source drop-down menu
- Set the X axis significance thresold to 10
- Set the Y axis significance thresold to 0.001
This gives 132 up-regulated and 159 down-regulated genes (Figure 9).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
We can now recreate these conditions in the ANOVA report filter.
- Click ANOVA report at the top of the screen to return to the ANOVA report
- Click FDR step up
- Set the FDR step up filter to Less than or equal to 0.001
- Click Fold change
- Set the Fold change filter to From -10 to 10
The filter should include 291 genes.
- Click to apply the filter and generate a filtered Feature list node
To visualize the results, we can generate a hierarchical clustering heat map.
- Click the second green Feature list
- Click Exploratory analysis in the task menu
- Click Hiearchical clustering
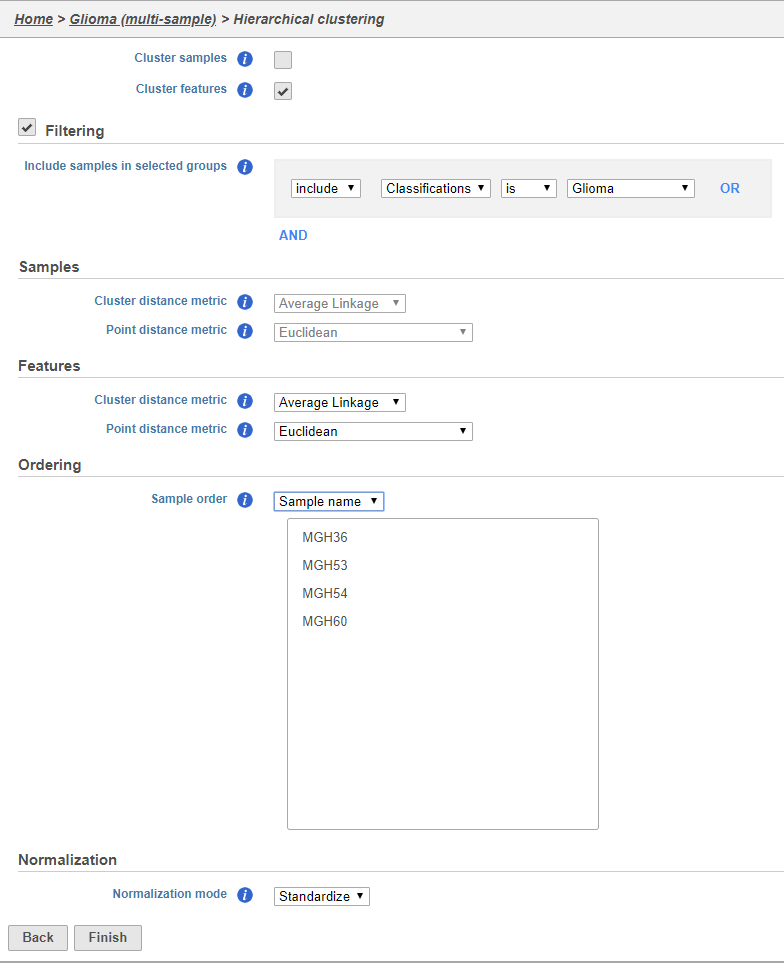
Using the hierarchical clustering options we can choose to include only cells from certain samples. We can also choose the order of cells on the heat map instead of clustering. Here, we will include only glioma cells and order the samples by sample ID (Figure 10).
- Uncheck Cluster samples
- Click Filtering and set the filter to include Classifications is Glioma
- Choose Sample name from the Sample order drop-down menu in the Ordering section
- Click Finish
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Double click the green Hierarchical clustering node to open the heat map
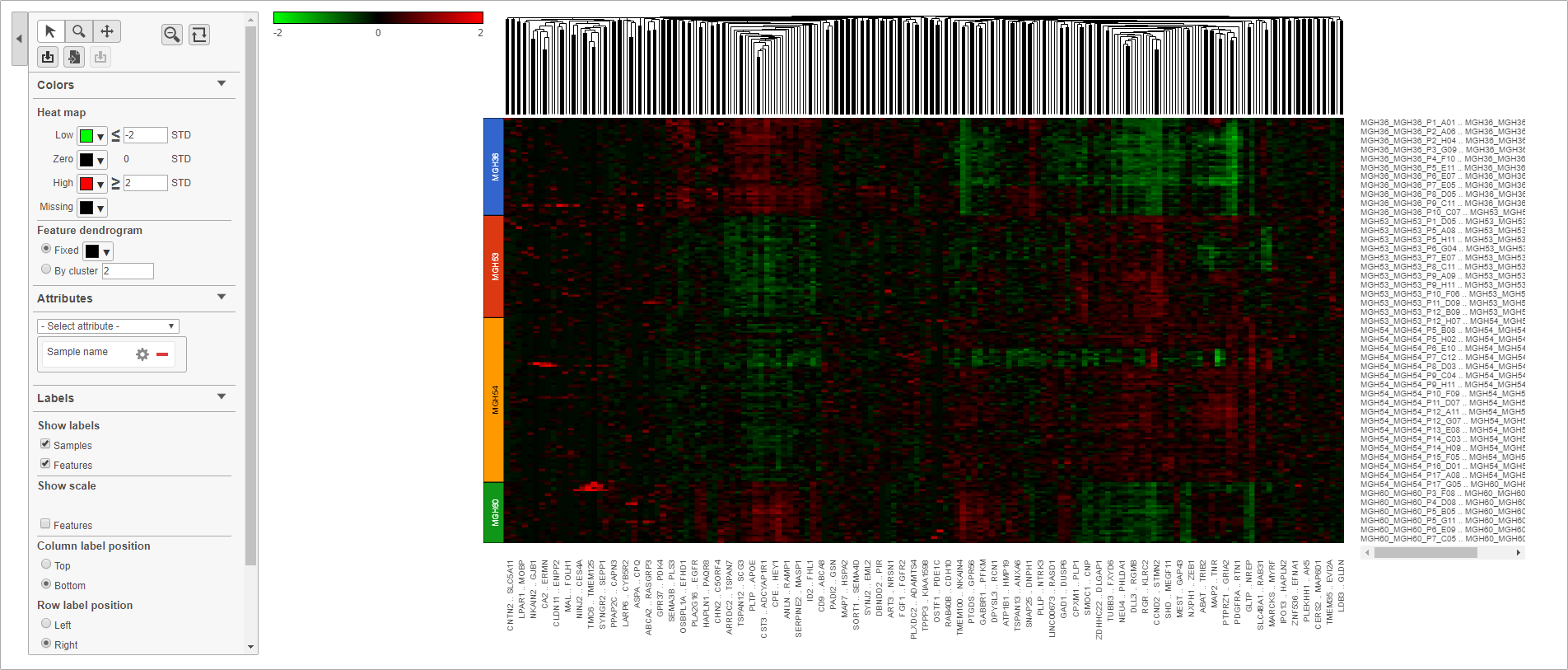
The heat map will appear black at first; the range from red to green with a black midpoint is set very wide because of a few outlier cells. We can adjust the range to make more subtle differences visible.
- Set Low to -2
- Set High to 2
The heat map now shows clear patterns of red and green.
- Select Sample name from the Attributes drop-down menu
Cells are now labeled with their sample name. Interestingly, samples show characteristic patterns of expression (Figure 11).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click Glioma (multi-sample) to return to the pipeline view
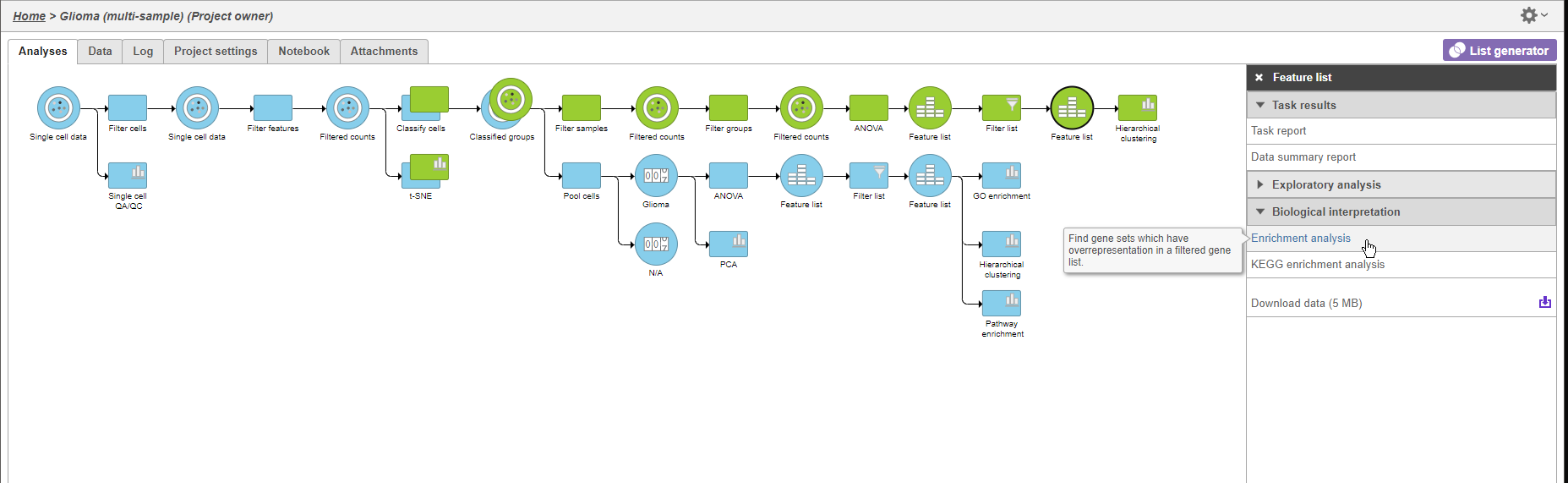
We can use GO enrichment to futher characterize the differences between glioma and oligodendrocyte cells.
- Click the second green Feature list node
- Click Biological interpretation in the task menu
- Click Enrichment anlaysis (Figure 12)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Choose Homo sapiens (human) - hg38 from the Assembly drop-down menu
- Select Finish to continue with the most recentgene set
A GO enrichment node will be added to the pipeline view (Figure 13).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
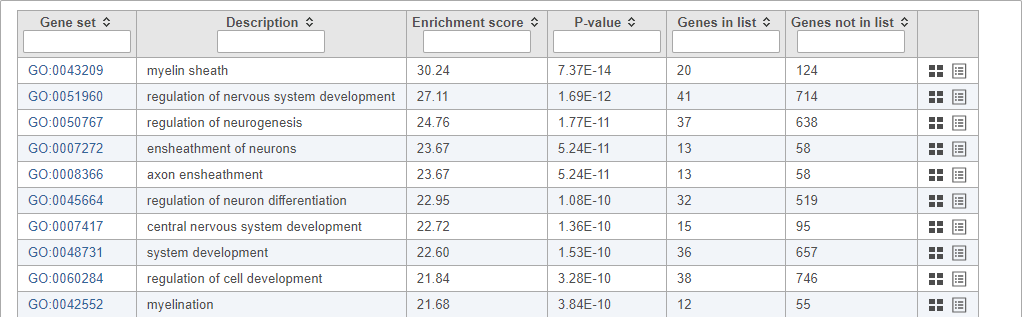
- Double-click the green GO enrichment task node to open the task report
Top GO terms in the enrichment report include "myelin sheath", "ensheathment of neurons", and "axon ensheathment" (Figure 14), which corresponds well with the role of oligodendrocytes in creating the myelin sheath that supports and protect axons in the central nervous system.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|