Page History
...
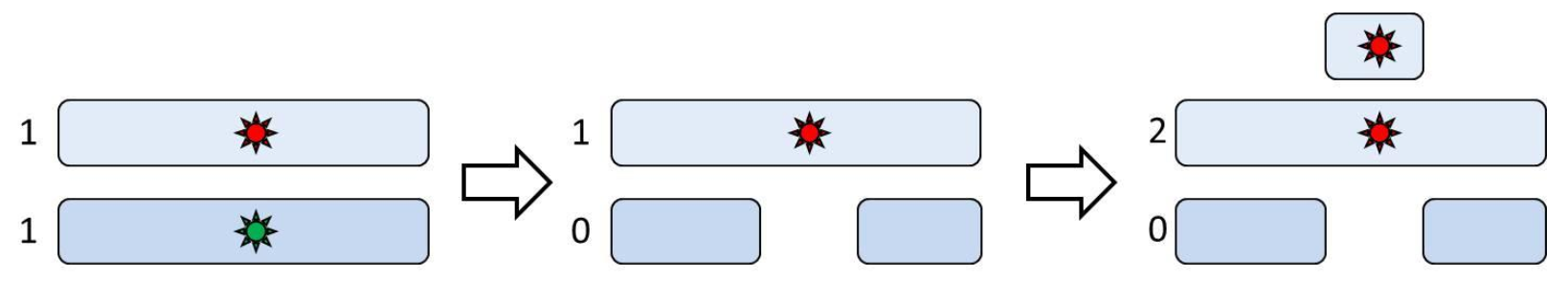
Although copy number analysis is a powerful tool for studying genomic aberrations, it lacks the capacity capability to detect genotypic changes which that are copy-neutral. If we consider For example, loss of heterozygosity (LOH) , this event may can involve a change in copy number or be copy-neutral. In the former case, LOH could be caused by a hemizygous deletion in which one allele is lost and the other allele remains present (Figure 1, middle panel). That This type of LOH can be recognized not only by copy number analysis or SNP-genotyping, but by copy-number analysis as well. However, in the latter case, an allele may is lost initially, but the a subsequent amplification of the remaining copy creates a copy-neutral LOH (Figure 1, right panel), also known as uniparental disomy. Different mechanisms have been described to create . This copy-neutral LOH in meiosis and mitosis; the common feature is that copy-neutral LOH can only be detected when copy number is studied in combination with SNP genotype. Please note that irrespective of the preservation of total number of copies, the biological effect is still important as recessive mutations would no longer be masked by their dominant normal counterparts.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
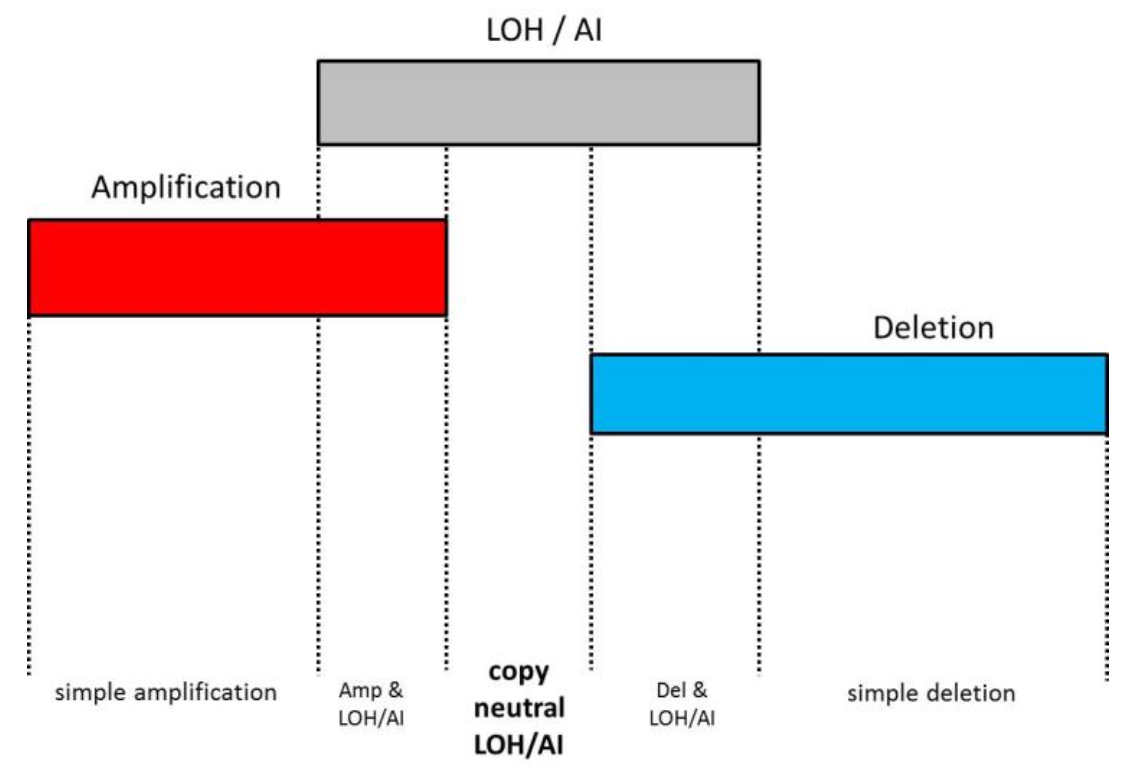
A solution for detection of copyCopy-neutral events is the integration of can be detect by combining the copy number workflow with the LOH workflow or the use of allelic imbalance (AI) under the Allele-Specific Copy Number (AsCN) workflow to detect allelic imbalance (AI) (advantages of AI AsCN over LOH are discussed below). With this approachthese approaches, the copy number data are supplemented with SNP genotyping data (currently available with Affymetrix® and Illumina® arrays) to label the genomic regions in the following fashion: as amplification without LOH/AI, amplification with LOH/AI, deletion without LOH/AI, deletion with LOH/AI, copy-neutral LOH/AI (Figure 2). The last category, copy-neutral LOH/AI, is the added value of the workflow integration.
A further consideration is that the correct interpretation of currently available algorithms for LOH An important consideration when choosing between LOH and AsCN analysis is that LOH analysis in the context of cancer has been proven complex and difficult because cancer cells frequently deviate from the diploid state , and tumor specimens samples often contain a significant proportion of normal cells. For instance, it has been shown that as many normal cells. As the proportion of tumor cells in a sample decreases and approaches 50% or less, the capacity to detect the LOH diminishes (Yamamoto et al., Am J Hum Gen 2007). Moreover, the Additionally, in cases where only one of two alleles is amplified, LOH genotyping algorithms fail to call a heterozygote SNP accordingly , resulting in a situation when only one of two alleles gets amplified (e.g. 3×A and 1×B): a false false-positive LOH call can be the consequence.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
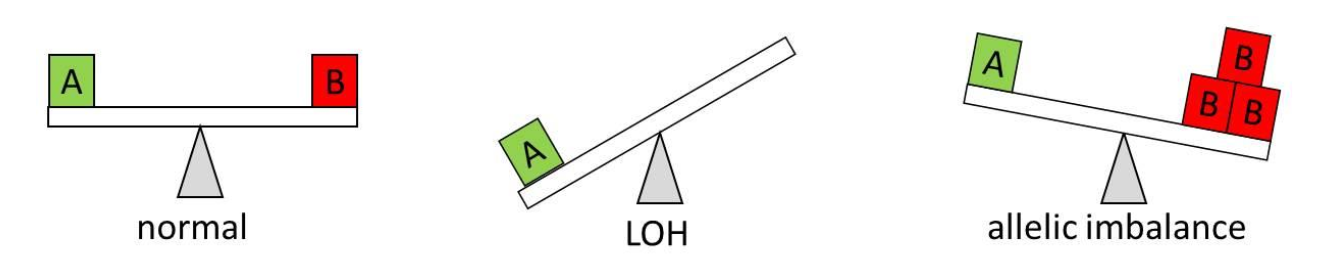
AsCN analysis, on the other hand, is a method that enables reliable detection of allelic imbalance in tumor samples even in the presence of large proportions of normal cells. Unlike LOH, it does not require a large set of normal reference samples. For a heterozygous SNP (only those are informative), a balance is expected between the two alleles (1×A and 1×B, or 1:1 ratio). The AsCN algorithm provides an estimated number of copies of each allele and therefore enables the detection of allelic imbalance even in cases when alleles are amplified or deleted (e.g. 3×A and 1×B). Moreover, LOH can be considered a special case of AI (e.g., 1×A, B allele deleted) (Figure 3). Therefore, due to its improved robustness, AsCN may be a preferred application in tumor-focused applicationsAsCN should be the preferred workflow for tumor samples.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...