Page History
...
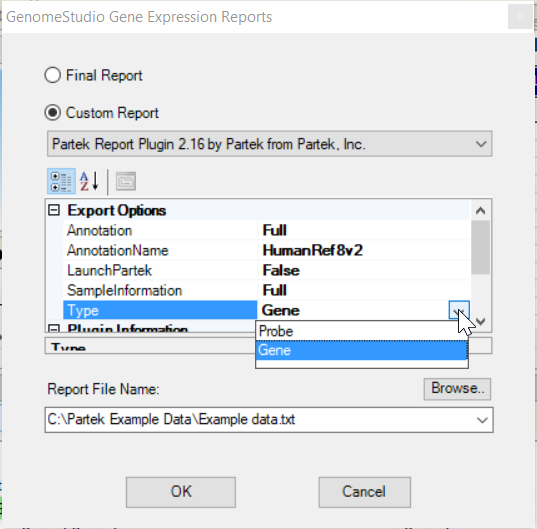
- Choose Analysis > Reports... from the main menu
- Select Custom Report and choose Partek Report Plug-in from the drop-down list
- Specify AnnotationName, remove the <>do NOT include <> in the name, you can use the chip namesame name as the .bgx file you imported the ddata with, or a unique name to your dataset
- Choose Type by clicking on the cell, default is can be gene level
- Leave all the others as default value (Figure 2)
- Specify the report file name and click OK
...
- , we recoommend to put the exported files in their own folder, which allows you to move thefolder instead of all the files individually.
- Click OK
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Open project in Partek Genomics Suite
To open the report, launch Partek Genomics Suite, choose File > Open Project, browse to the .ppj file to open. In the Gene expression workflow, you can proceed add sample attribute step.
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
Overview
Content Tools