Page History
...
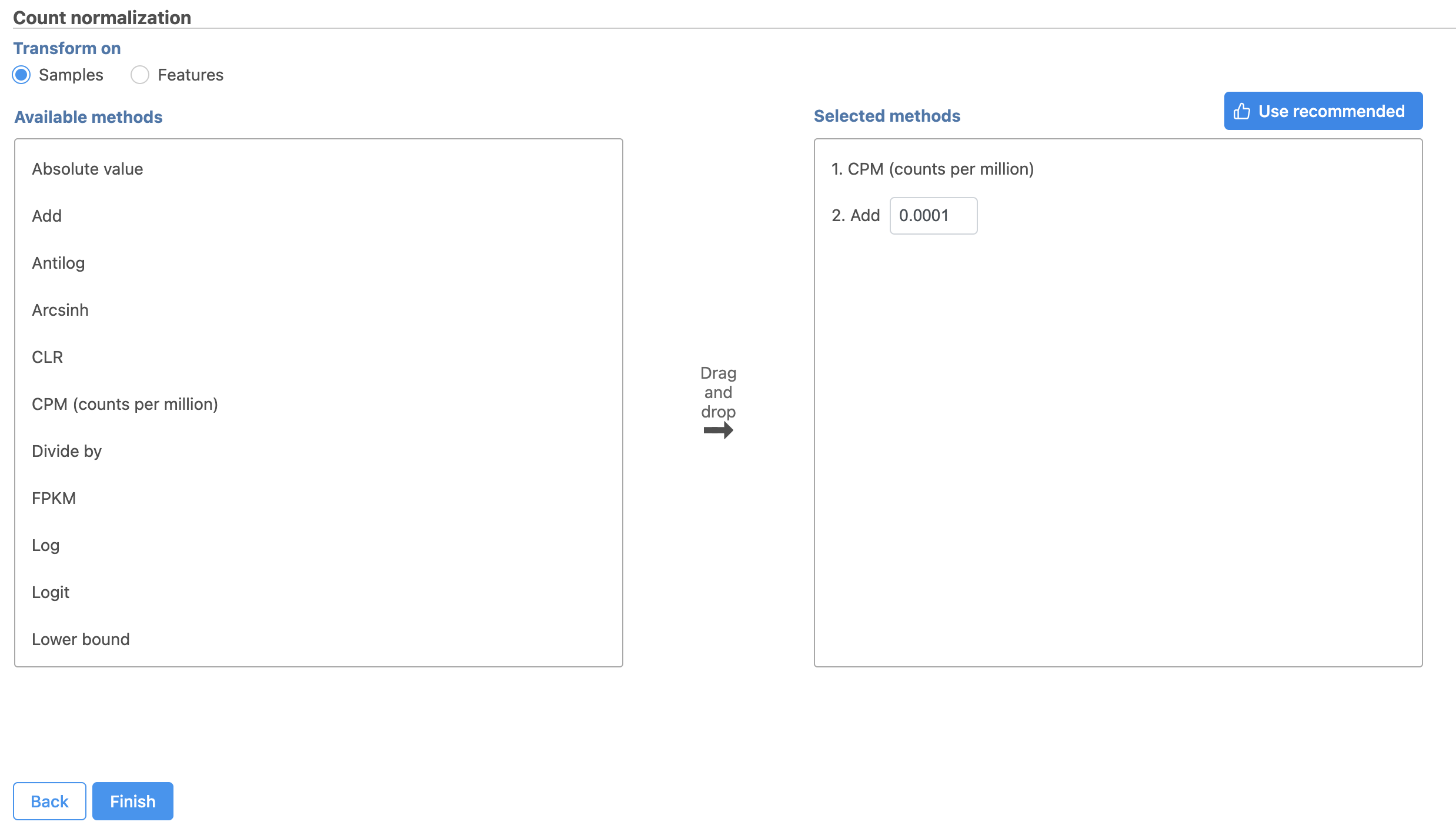
For this tutorial, we will use the recommended default normalization settings.
- Select
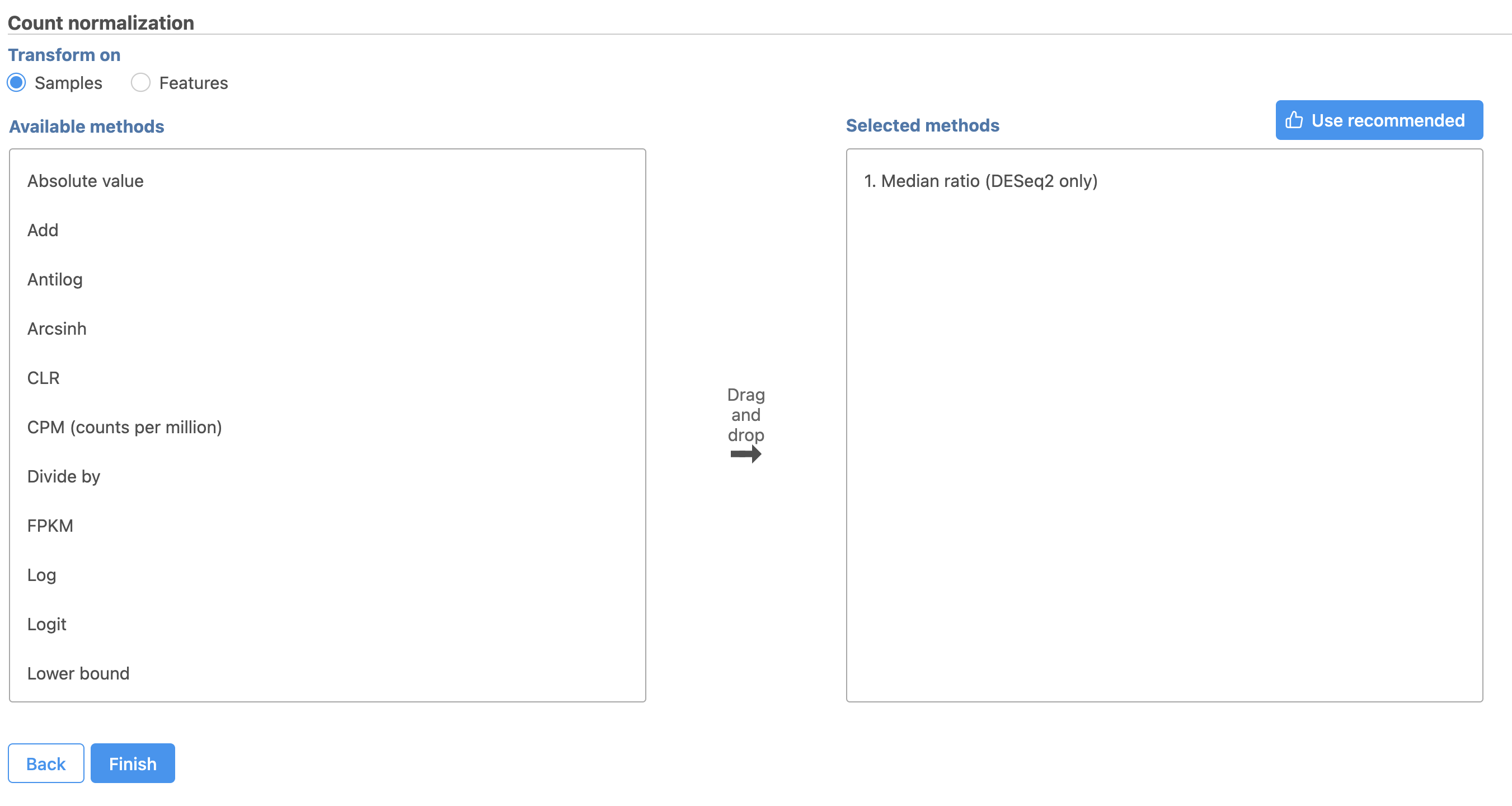
This adds CPM (counts per million) and Add 0.0001 to the Normalization order panel (adds the Median ratio normalization method, which is suitable for performing differential expression analysis using DESeq2 (Figure 3). Normalization steps are performed in descending order
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
CPM (counts per million) normalizes read counts for each gene by the total count of the sample. This accounts for differences in total read counts between samples.
Add 0.0001 adds 0.0001 to the normalized read count of every gene. This prevents the read count data from having any 0 values. Values of 0 would prevent the gene specific analysis algorithm we will use for differential expression analysis from performing the necessary log transformation.
- Click Finish to perform normalization
...