Page History
...
With attributes added, we can begin building our pipeline.
- Select Click the Analyses tab
In Partek Flowthe Analysis tab, data are represented as circles, termed data nodes. One data node, Unaligned readsmRNA, should be visible in the analysis Analysis tab (Figure 1).
- Select the Unaligned reads data node
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click the mRNA node
Clicking a data node brings up a the context-sensitive task menu that lists with tasks that can be performed on the data node (Figure 2).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Pre-alignment QA/QC to assess assesses the quality of the unaligned reads and will help us determine whether trimming or filtering is necessary.
- Select Click Pre-alignment QA/QC from in the QA/QC section of the task menu
- Select Click Finish to run the task with default settings
Selecting Running a task creates a task node, e.g. the blue rectangle labeled Pre-alignment QA/QC (Figure 3). Selecting the task node will let us view the task report and the task details.
...
, which contains details on the task and a report.. While tasks have been queued or are in progress they have a lighter color. Any output nodes that the task will generate are also displayed in a lighter color until the task completes. Once the task begins running, a progress bar is displayed on the task node.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
- Click the Pre-alignment QA/QC task node
The context-sensitive task menu (Figure 4) shows the option to view the task Task report and the task Task details. You can also access a task report by double-clicking on a task node.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click Task report
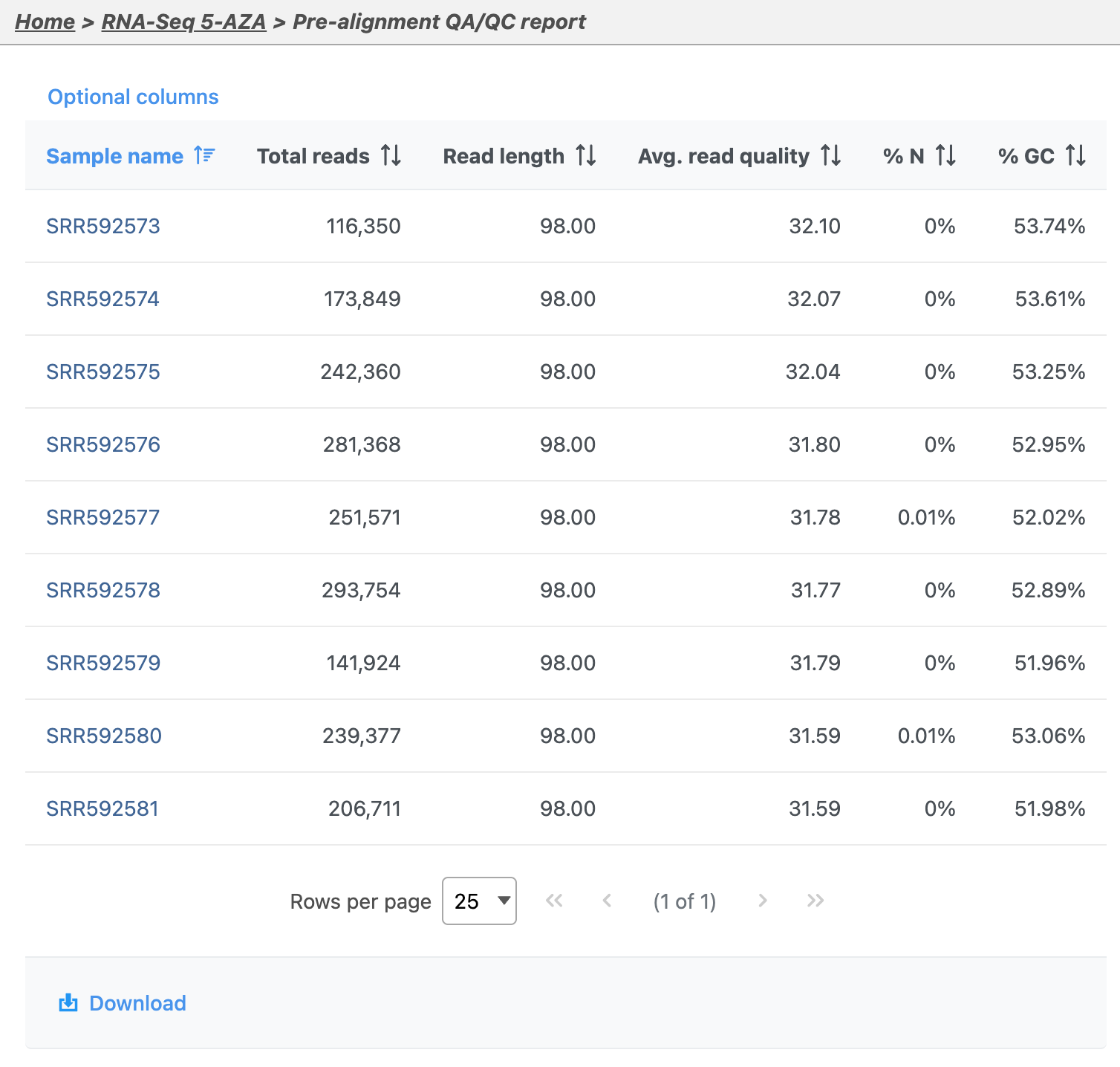
Pre-aligment QA/QC provides information about the sequencing quality of unaligned reads (Figure 5). Both project level summaries and sample-level summaries are provided. To view sample-level summaries, select a sample from the data table in the report.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click sample SSR592573 in the data table of the report to open its sample-level report
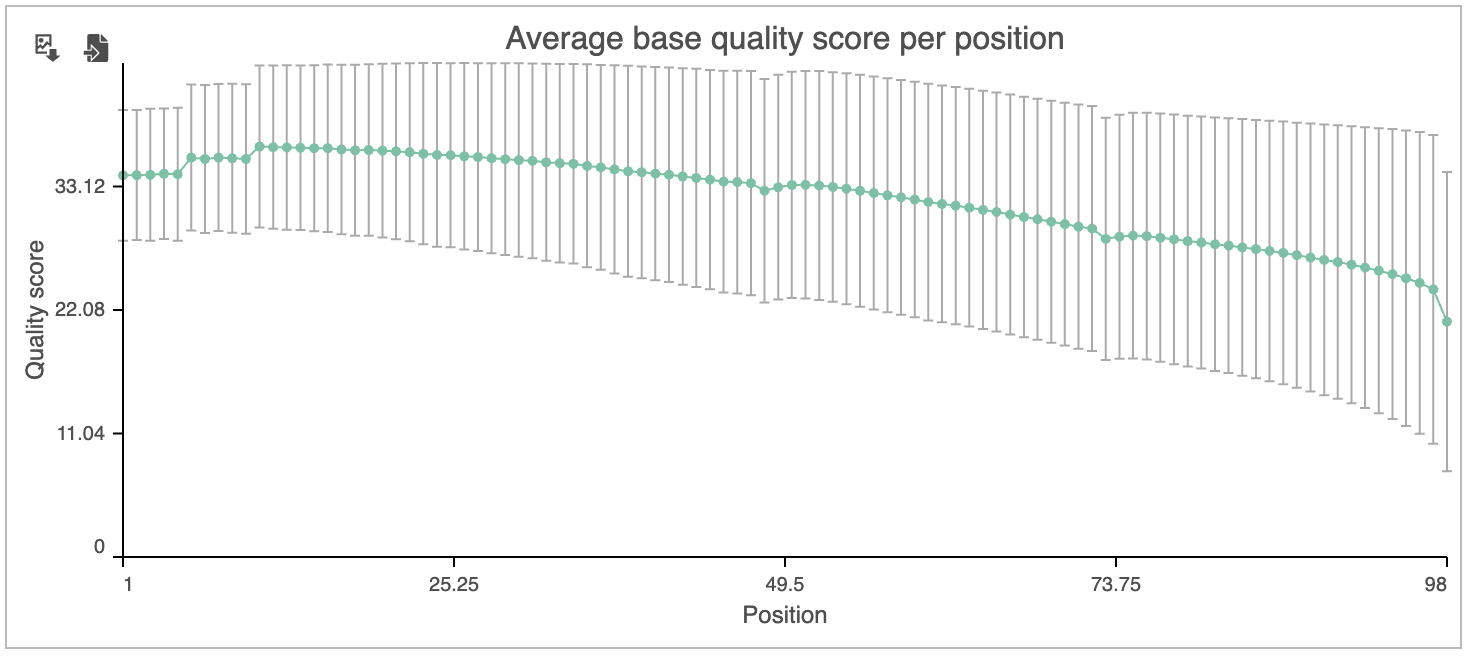
The Average base quality score per position graph in the upper right-hand panel (Figure 6) gives the average Phred score for each position in the reads.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A Phred score is a measure of base call accuracy with a higher score indicating greater accuracy.
Phred Quality Score | Probability of incorrect base call | Base call accuracy |
|---|---|---|
20 | 1 in 100 | 99% |
30 | 1 in 1000 | 99.9% |
40 | 1 in 10,000 | 99.99% |
50 | 1 in 100,000 | 99.999% |
By convention, a score above 20 is considered adequate. As we you can see, the standard error bars in the graph show that some reads have quality scores below 20 for some of their base pair calls .
Similarly, the average base quality score per read graph (Figure) shows that some reads have average Phred quality scores below 20.
near the 3' end.
Based on the results of Pre-alignment QA/QC, while most of the reads are high quality, we will need to perform read trimming and filtering. For more information about the information included in the task report, please see the Pre-aligment alignment QA/QC User Guide user guide.
- Click RNA-Seq 5-AZA to return to the Analyses tab
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...