Page History
...
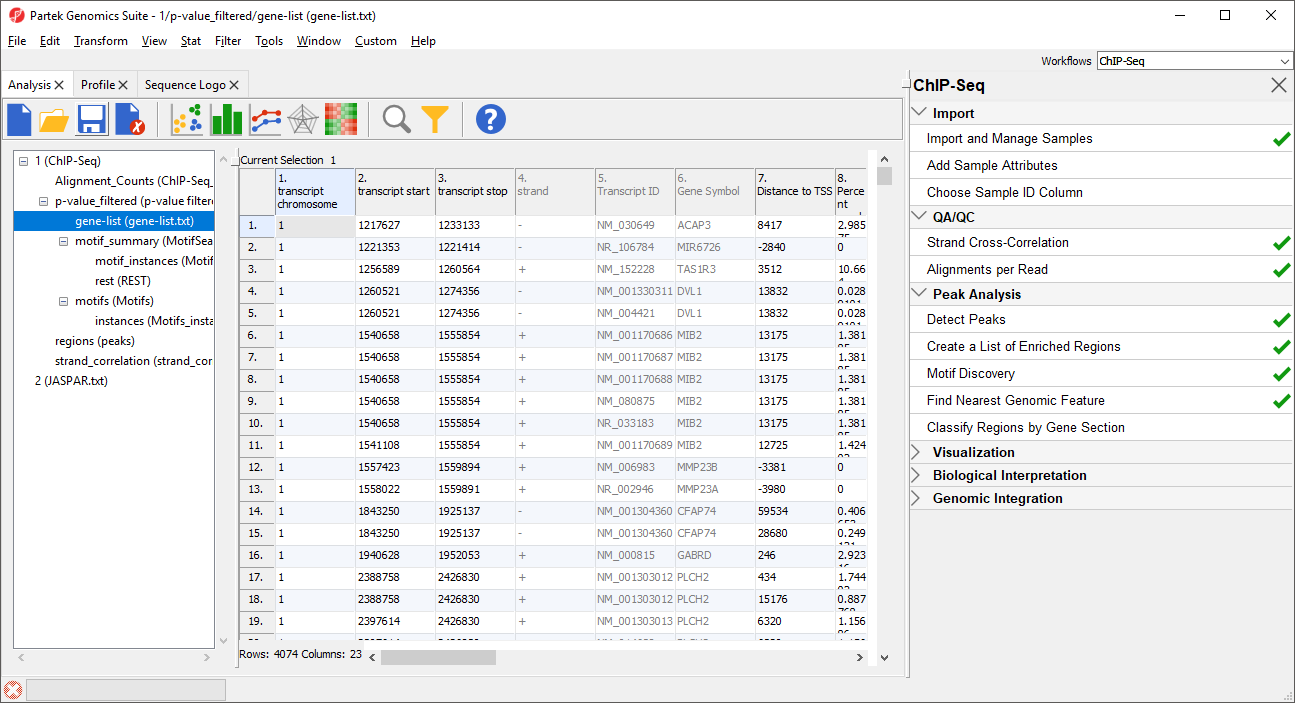
- Select p-value_filtered from the spreadsheet tree
- Select Find nearest genomic features Nearest Genomic Feature from the Peak Analysis section of the ChIP-Seq workflow
...
With this dialog, you can specify the reference database. Here
- Select RefSeq Transcripts 81 -
...
- 2017-
...
- 08-02 or your preferred annotation database
The promoter region can also be defined. The default settings are appropriate in this case.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Column 1. transcript chromosome gives the chromosome location of transcript
...
- Select p-value_filtered from the spreadsheet tree
- Select Classify regions by gene selection from the Peak Analysis section of the ChIP-Seq workflow
The Output Overlapping Features dialog will open.
- Select RefSeq Transcripts 81 - 2017-08-02 or your preferred annotation database
The promoter region can also be defined. The default settings are appropriate in this case. The results can be further configured to give one result per detected region or one result per genomic feature. The default setting, one result per detected region, is appropriate in this case.
- Select OK
A new spreadsheet, gene-classification will be generated (Figure 3).
...
Overview
Content Tools