Page History
...
Data for this tutorial can be downloaded from the Partek website using this link - ChIP-Seq tutorial data. To follow this tutorial, download the two .bam files and unzip them on your local computer using 7-zip, WinRAR, or a similar program.
- Store the 2 two.bam files at C:\Partek Training Data\ChIP-Seq or to a directory of your choosing. We recommend creating a dedicated folder for the tutorial on a local drive.
- Select ChIP-Seq from the Workflows drop-down menu (Figure 1)
...
- Set Output file to ChIP-Seq
- Set Species to Homo sapiens using the drop-down menu
- Set Genome build tohg18 to hg18 using the drop-down menu
- Select OK
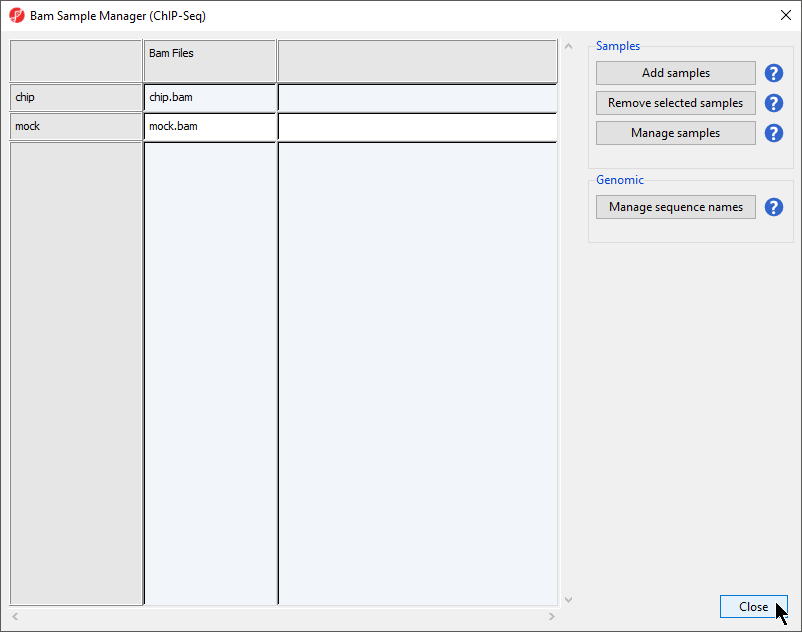
The Bam Smaples Samples Manager dialog will open (Figure 4). This dialog can be used to add samples to the project (Add samples), remove samples (Remove samples), to associate multiple files with particular samples (Manage samples), and to map the chromosome names form from the input files to the association files (Manage sequence names).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Close
You may see the The Sort bam files dialog . If so, select OK. The will open. Sorting is necessary for all imported .bam files, but you can choose to hide this hint in the future by selecting Please don't show me this hint again.
- Select OK
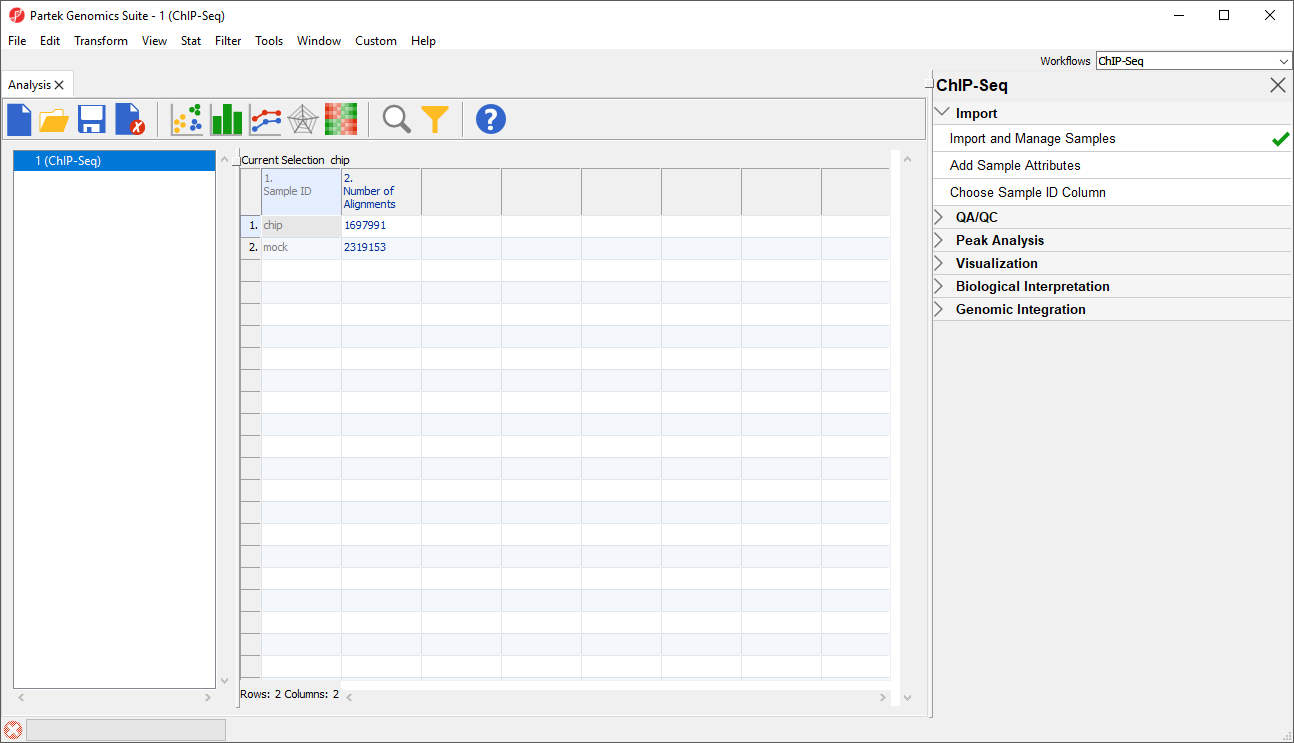
The imported spreadsheet will open after import completes (Figure 5). In this spreadsheet, samples are rows while the .bam files are sorted. Progress in sorting will be displayed on the progress bar in the lower left-hand corner of the Partek Genomics Suite window. Once sorting has completed, there will be samples on rows with the sample names in column 1. Sample ID and the number of aligned reads is listed in column 2reads mapped to the reference genome for each sample in column 2. Number of Alignmentsallignments (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|