Page History
To analyze differences in methylation between our experimental groups, we need to create a list of deferentially methylated loci.
- Select Create Marker List from the Analysis section of the Illumina BeadArray Methylation workflow
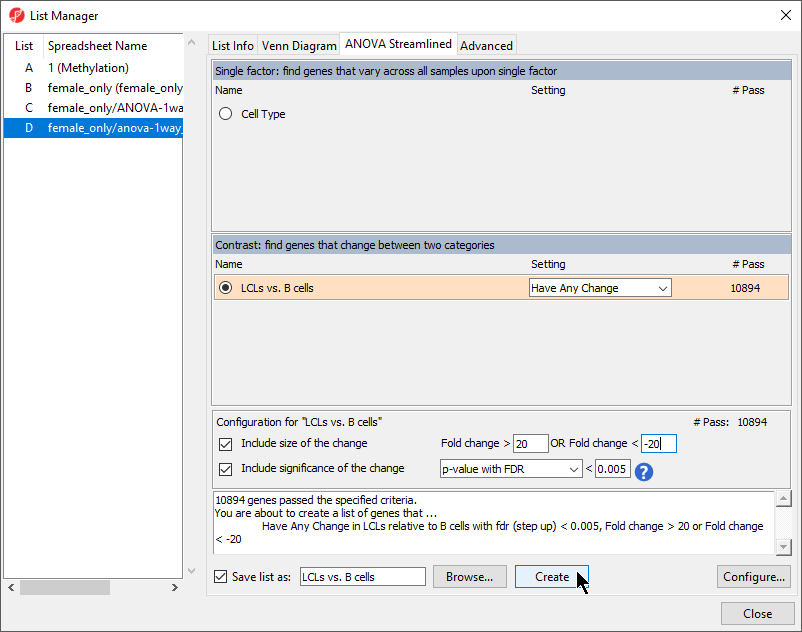
- Select anova-1way_autosomal (autosomal) as the source spreadsheet from the left-hand panel
- Select LCLs vs. B cells from the Contrast: find genes that change between two categories section of the ANOVA Streamlined tab of the List Manager dialog
- Set Fold change > to 20
- Set OR Fold change < to -20
- Set p-value with FDR to 0.005
- Set file name as LCLs vs. B cells
- Leave the rest of the option set to defaults (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Create
- Select Close to exit the List Manager dialog
The new spreadsheet LCLs vs. B cells (LCLs vs. B cells) will open in the Analysis tab (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
Overview
Content Tools