Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Hierarchical Clustering
The gene list in spreadsheet Down_Syndrome_vs_Normal (A) can be used for hierarchical clustering to visualize patterns in the data.
...
For more information on the methods used for clustering, you can refer to Chapter 8: Hierarchical & Partitioning Clustering in Help > User’s Manual. For a tutorial on configuring the clustering plot, please refer to .
Adding Gene Annotations
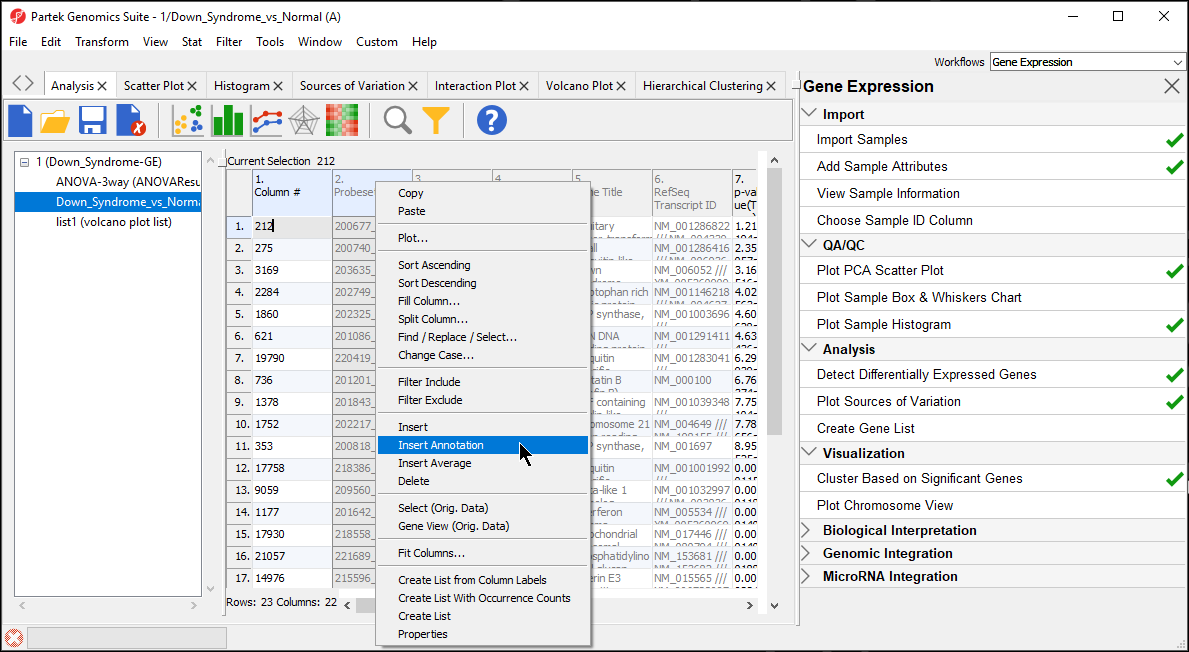
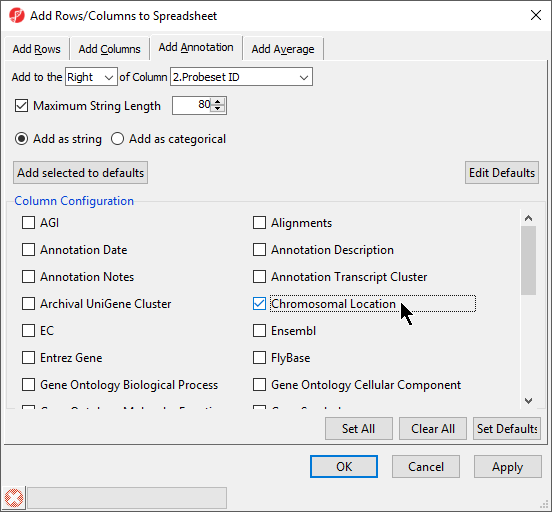
During data importation, the GeneChip annotation file was linked to the imported data. This linked annotation information can be added as new columns to the ANOVA or gene list spreadsheets. For example, we can add additional annotation to the gene list we created from the ANOVA results as follows:
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Interestingly, of the 23 genes of the Down_Syndrome_vs_Normal (A) spreadsheet, 20 genes are located on chromosome 21. This suggests that the gene expression changes associated with Down syndrome observed in this study are primarily located on chromosome 21, not distributed throughout the genome, an important finding of this study.
To save changes to the spreadsheet, select the Save Active Spreadsheet icon ().
Hierarchical Clustering Analysis.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|