Page History
...
For Partek Genomics Suite to recognize an annotation spreadsheet, it must meet three several requirements. First, there must be a column header row in the annotation file. Second, there must be a column in the annotation file that matches the probe identifiers in your data spreadsheet. Third, any text field above the column header row must start with #. Fourth, the fields must be tab or comma delimited.
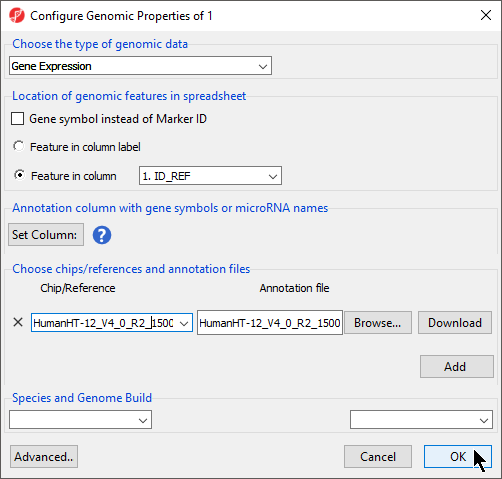
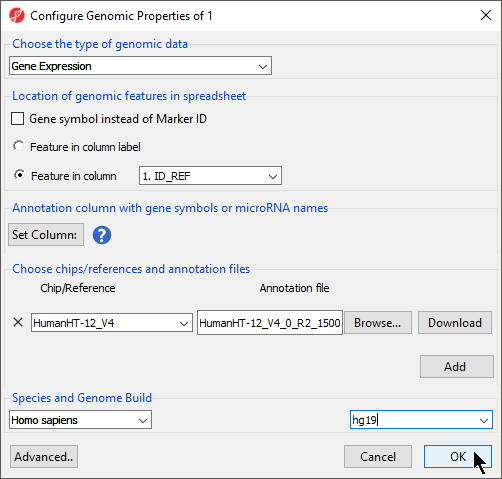
We will illustrate annotation using an imported .txt data files file from an Illumina HumanHT-12 v4.0 Gene Expression BeadChip array and the HumanHT-12 v4.0 Whole-Genome Manifest File (TXT Format) from Illumina.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select OK apply the annotation file to your data spreadsheet
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
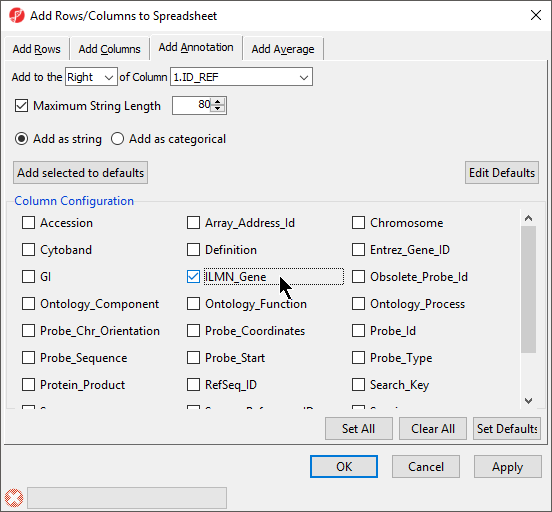
Annotation features can also be added to the Genome Viewer as tracks. See (chapter on genome viewer) for an introduction to adding tracks.
Building an Annotation File
If you are using an Illumina, Affymetrix, or an Agilent chip, you can obtain the appropriate annotation file Annotation files for most commercial arrays are available from the chip manufacturer. If you have a custom chip or want to use a customized annotation file, you can create an annotation file that will allow you to add annotations to your data spreadsheet and invoke a Probe Set HTML report. Your annotation file must meet the following criteria:
- The annotation file must have a column header (with a line which contains a column label for each field) column
- A column in the annotation file must correspond to the feature ID column of your data spreadsheet
- Any comments before the header must start with # or the header will not be recognized
- The fields of the annotation file must be tab or comma delimited
- A column in the annotation file must correspond to the feature ID column of your data spreadsheet
To invoke a genome view of your data, your annotation file must also have one or more columns that contain the genomic location in a format that Partek can recognize.
...