Page History
...
Partek Genomics Suite enables you to visualize each probe and compare the methylation between the groups at a single CpG site level. The visualization can be supplemented with a special annotation file which can be built using a UCSC annotation file as the basis.
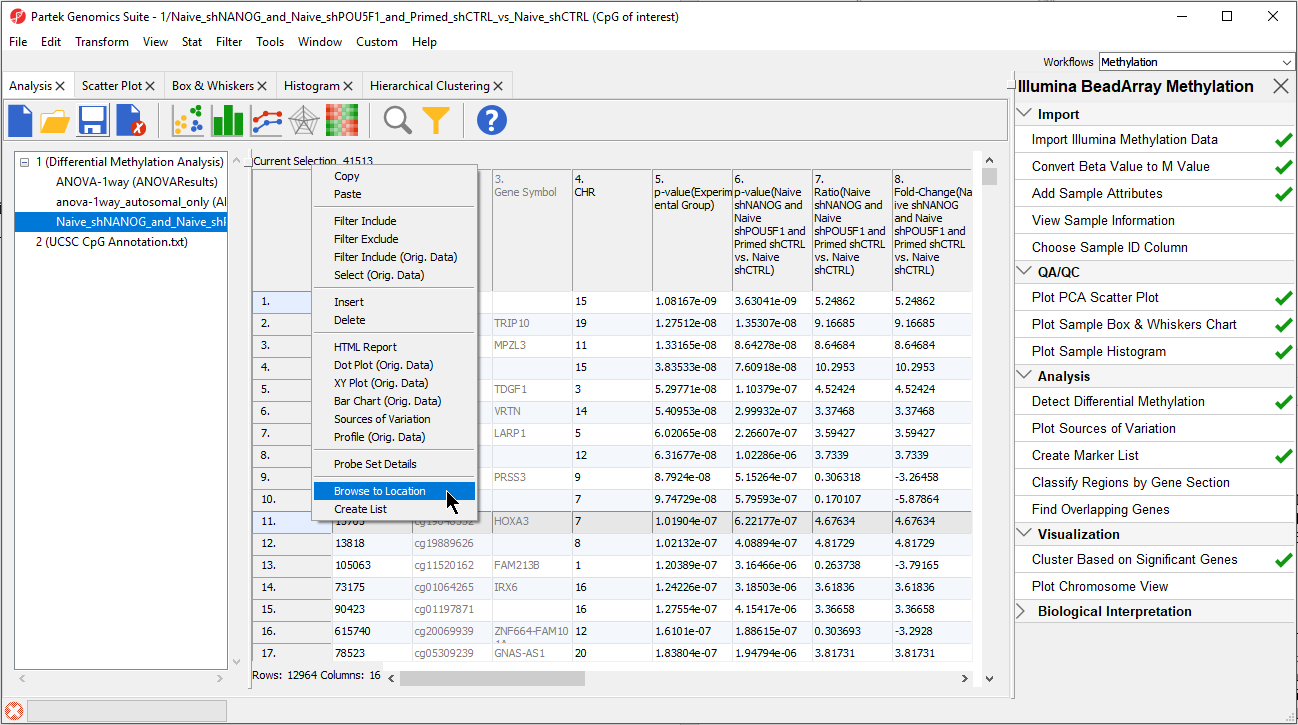
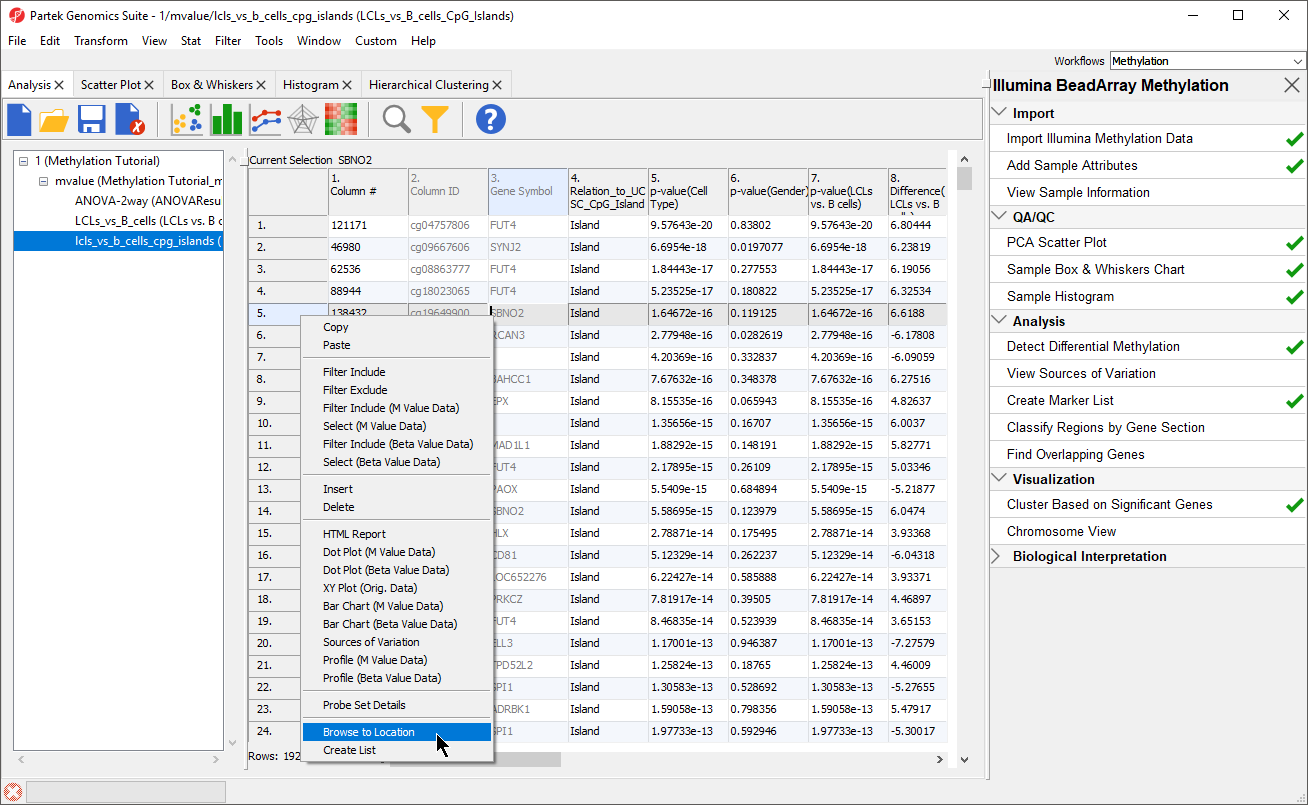
- Right click row 5. SBNO2 in the LCLs_vs_B_Cells_CpG_Islands spreadsheet
- Select Browse to Location from the pop-up menu
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Next, we can import the BED file into Partek Genomics Suite.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Adding UCSC CpG Annotations
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
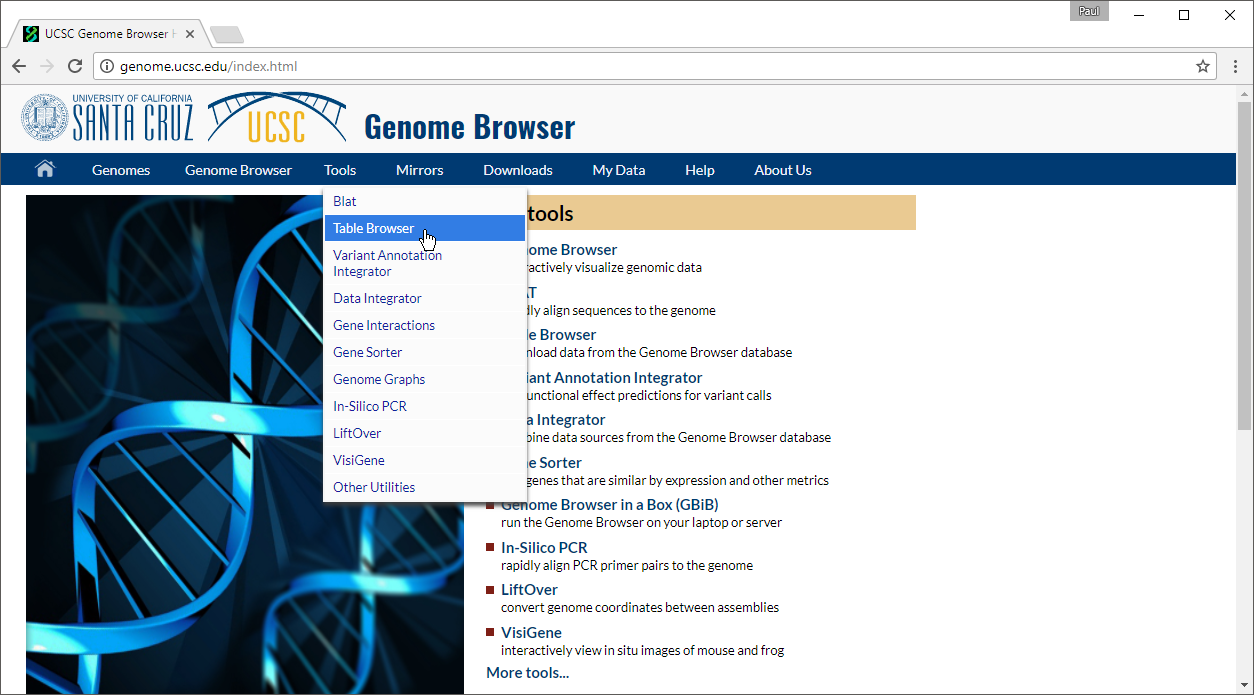
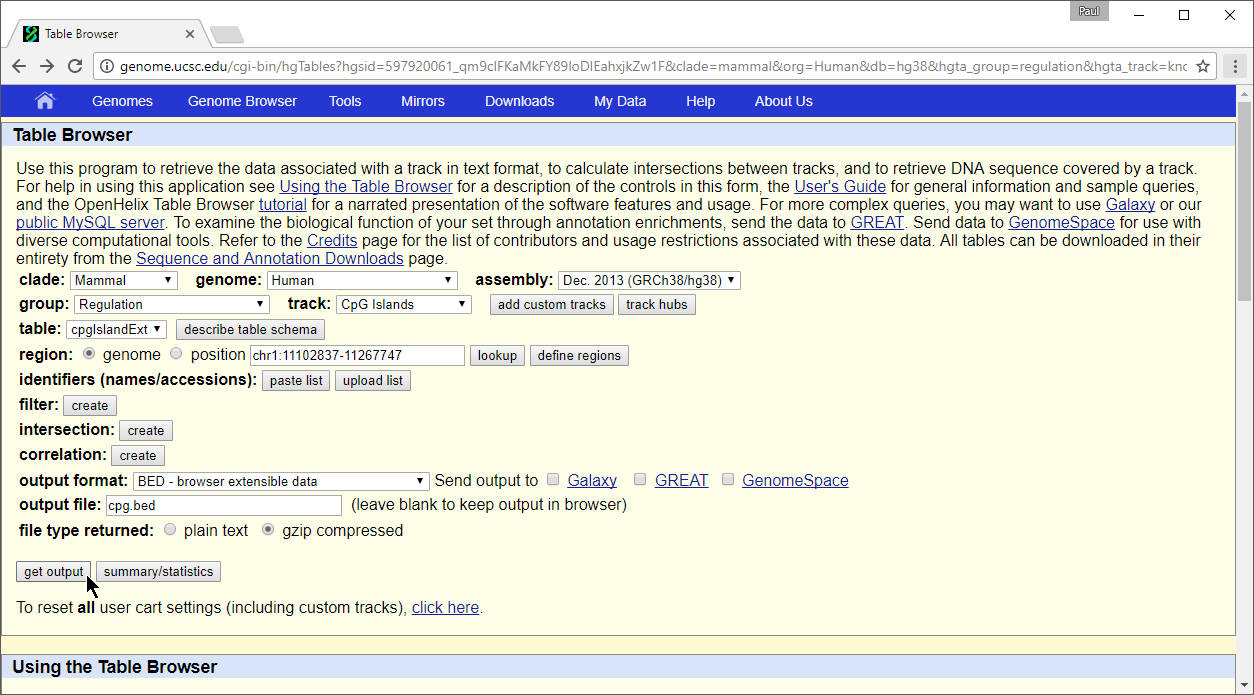
- Set group to Regulation
- Set track to CpG Islands
- Set table to cpgIslandExt
- Set output format to BED
- Set output file to cpg.bed
- Select get output
The Output cpgIslandExt as BED page will open.
- Select get BED to download a compressed folder containing the BED file
- Unzip the file using 7-Zip, WinRAR, or a similar program of your choice to a location you will be able to find
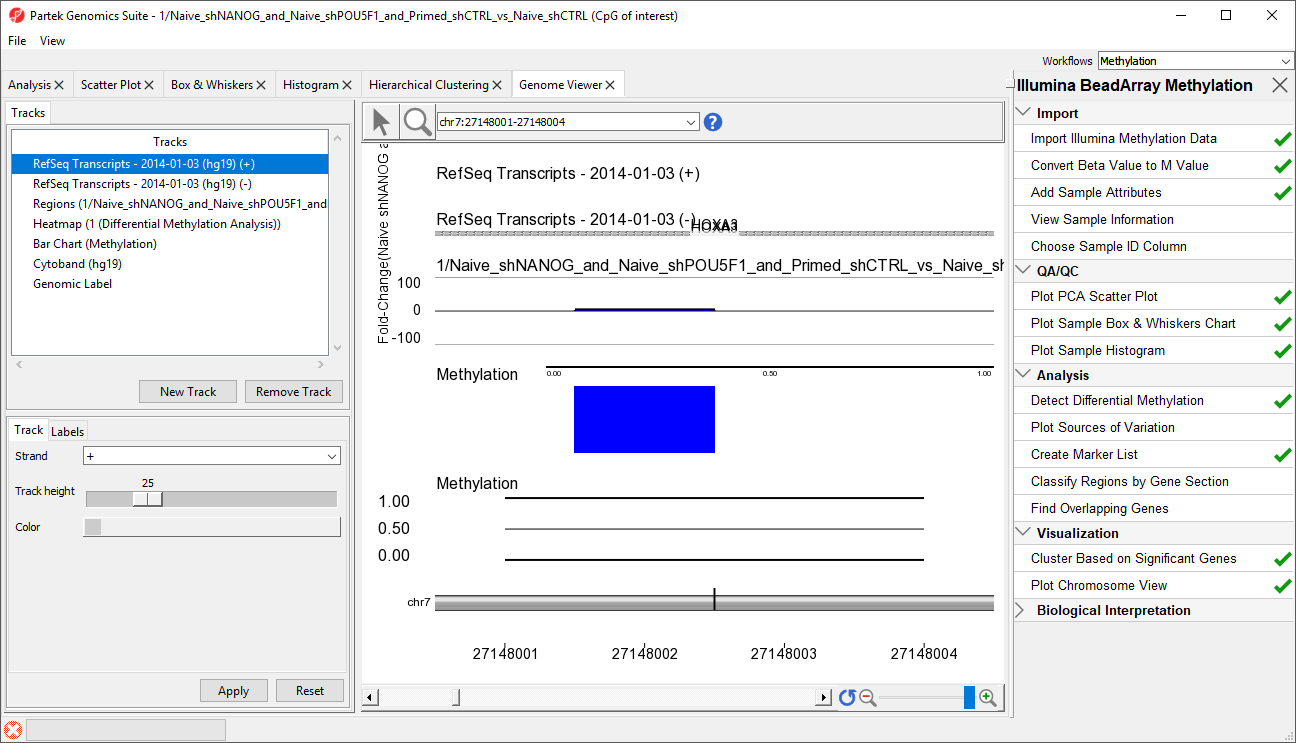
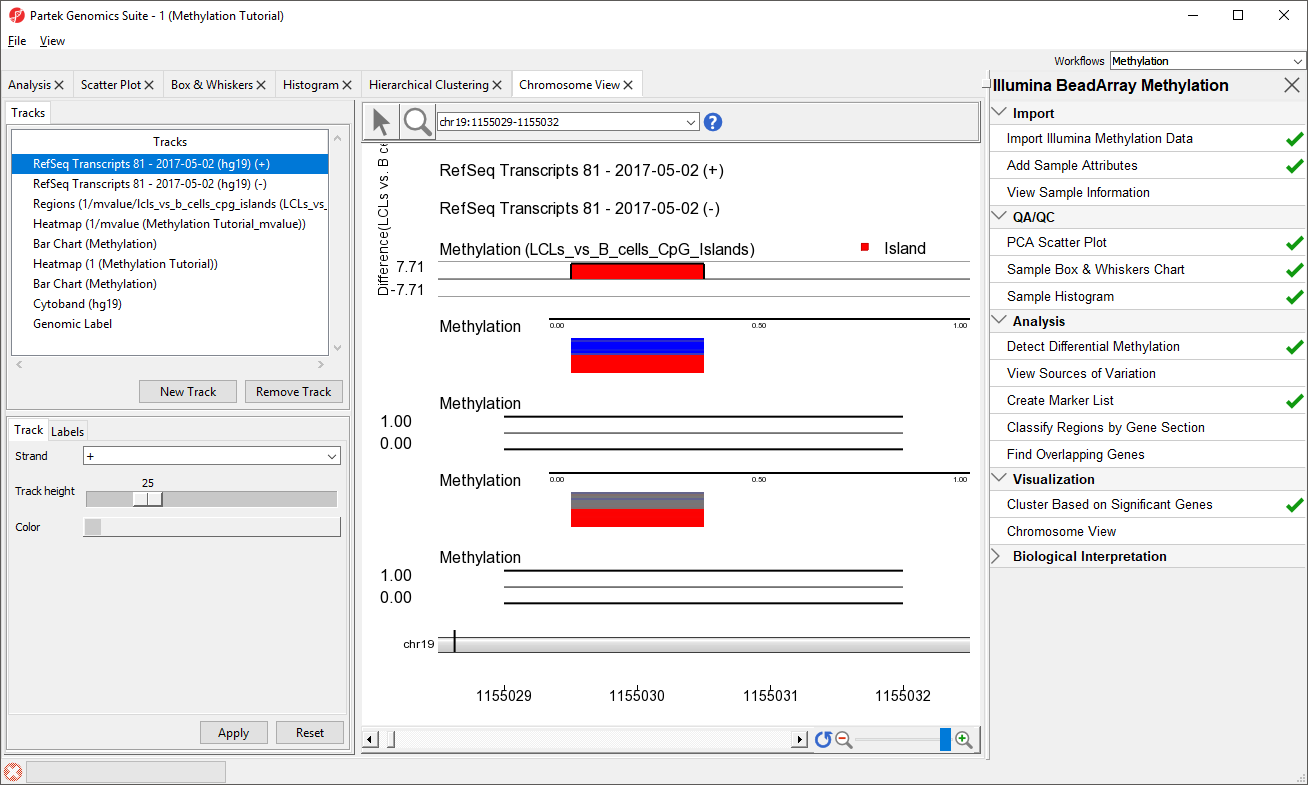
- RefSeq Transcripts 2017-05-02 (hg19) (+): transcripts coded by the positive strand
- RefSeq Transcripts 2017-05-02 (hg19) (-): transcripts coded by the negative strand
- Regions: by default, difference in methylation (M-value) between the groups
- Heatmap (1/mvalue): M values for all the samples
- Barchart (Methylation): methylation level in M value of the selected sample (to select a sample, click on a heat map)
- Heatmap (Methylation Tutorial): Beta values for all the samples
- Barchart (Methylation): methylation level in Beta value of the selected sample (to select a sample, click on a heat map)
- Cytoband: cytobands of the current chromosome
- Genomic Label: coordinates on the current chromosome
To modify a track, select it in the Tracks panel to bring up its configuration options panel below the Tracks panel. Let's modify a few tracks to improve our visualization of the data.
- Select the Regions track, opens to Profile tab
- Select Color tab
- Set Color bars by to Difference (LCLs vs. B cells) (Description)
- Select Apply to change
This will color regions by up or down methylated.
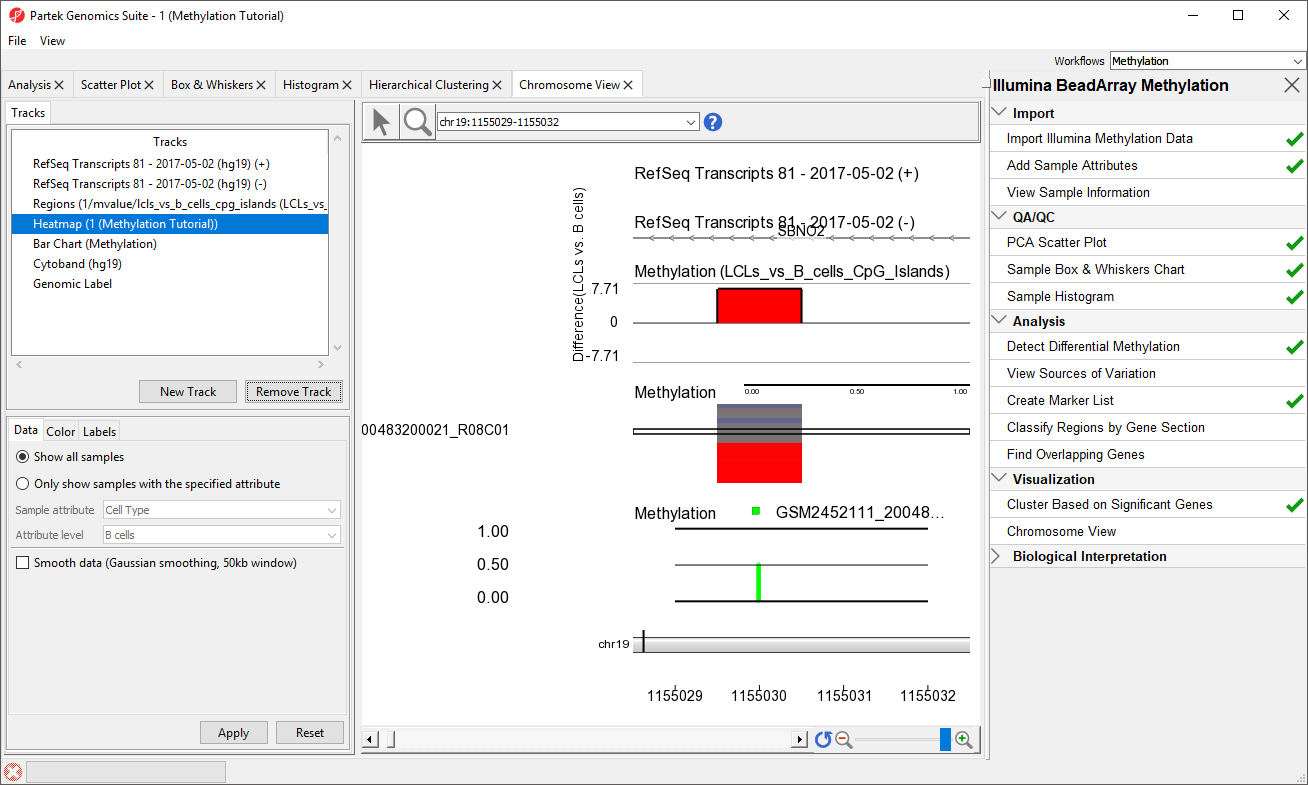
- Select the Heatmap (1/mvalue)
- Select Remove Track
- Select Bar Chart (Methylation) located directly below the Regions track
- Select Remove Track
We can now more clearly see the Difference in M values for the region in the Regions track, the heatmap of beta values in the Heatmap track, and the beta value for the loci of the selected sample in the Bar Chart track.
- Select a sample on the heatmap to view its beta value in the Bar Chart track (Figure 3)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
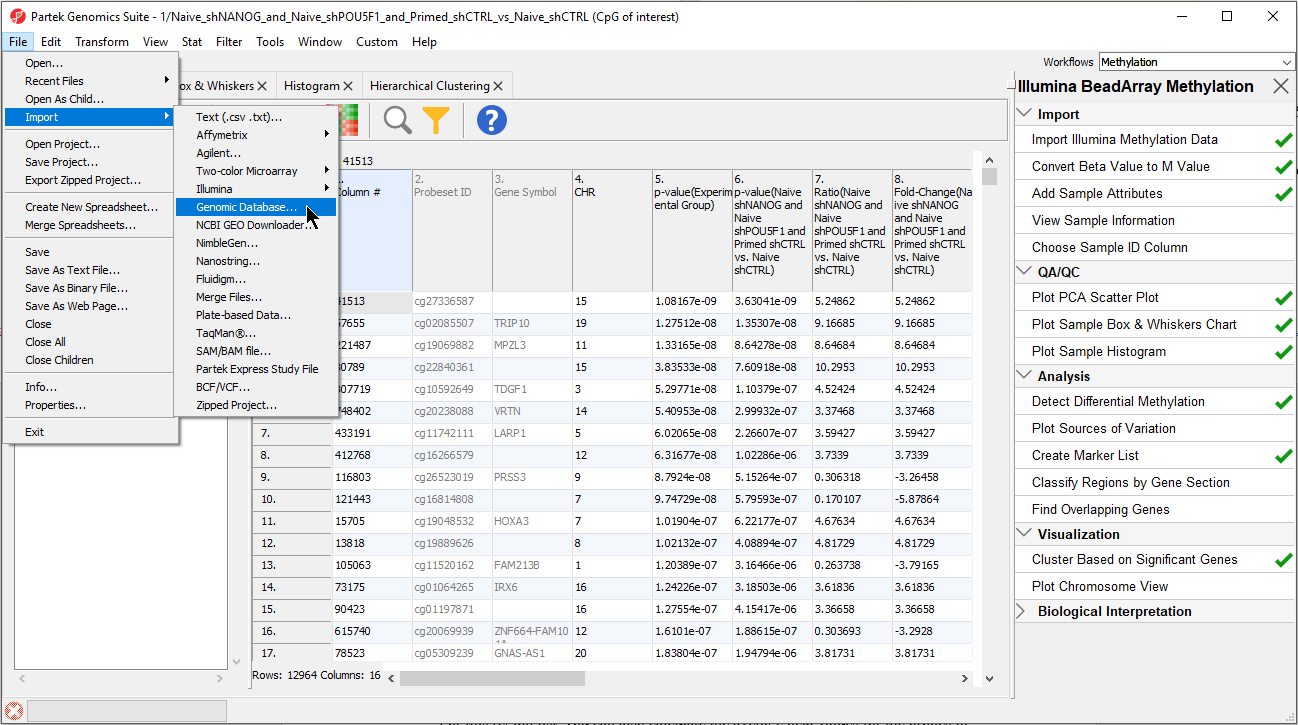
- Select the file cpg.bed
The BED file will open as a new spreadsheet (Figure 5).
- Change the spreadsheet name to UCSC CpG Island Annotation and save it
For this region list, you can also calculate the average beta values for the probes in each island per sample and detect differential methylated CpG islands regions. Detailed information on how to get average beta value for each CpG can be found from the Determining the average values for a region list section of Help>On-line Tutorials>User Guides> List.
| |
The New Track button allows new tracks to be added to the viewer, while the Remove Track button removes the selected track from the viewer. Tracks can be reordered by selecting a track in the Tracks panel and dragging it up or down to move it in the list. In the Chromosome View, select () for selection mode and () for navigation mode. In navigation mode, left-click and draw a box on any track to zoom in. All tracks are synced and will zoom together. Zooming can also be controlled using the interface in the lower right-hand corner of the tab (). View can be reset to the whole chromosome level using reset zoom (). Searching for a gene or transcript in the position box will also zoom directly to its location.
The available tracks can be supplemented with a special annotation file that can be built using a UCSC annotation file as the basis. Building and viewing the UCSC annotation file is available as an optional section of the tutorial, Optional: Add UCSC CpG island annotations.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|