Join us for an event September 26!
How to Streamline RNA-Seq analysis and increase productivity—point, click, and done
Partek Genomics Suite software can view annotation .BED files as tracks in the Genome Viewer. We can add a CpG islands track to the Genome Viewer using the UCSC Genome Browser CpG islands annotation.
- Go to UCSC Genome Browser
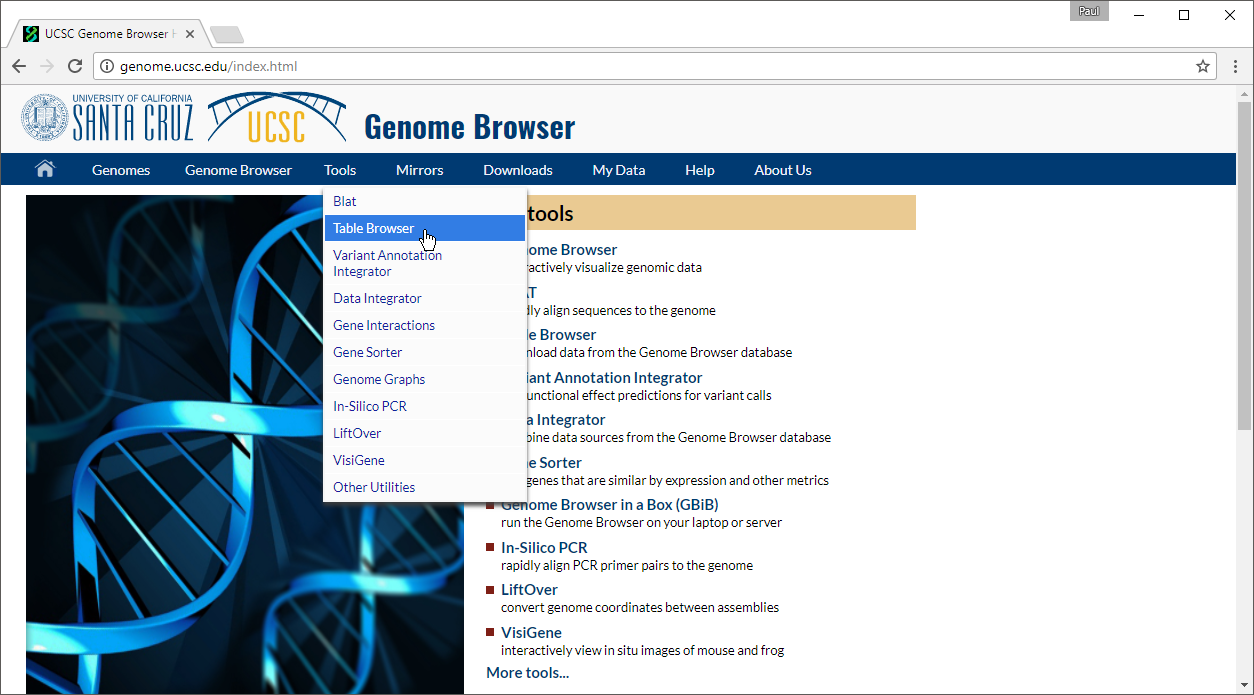
- Select Table Browser under Tools in the main command bar of the webpage (Figure 1)
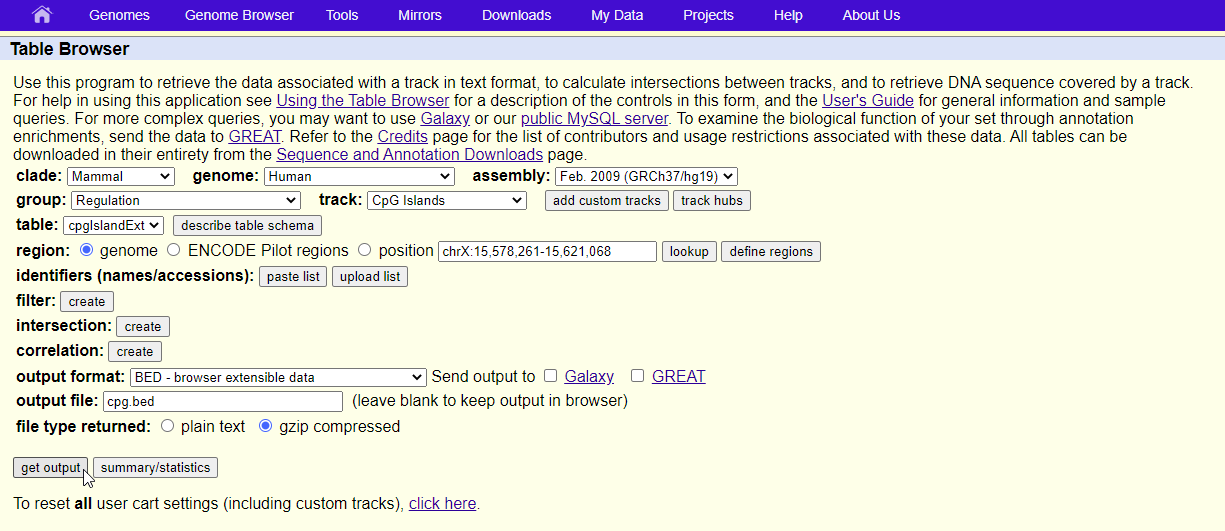
- Configure the Table Browser page as shown (Figure 2)
- Set assembly to Feb. 2009 (GRCh37/hg19)
- Set group to Regulation
- Set track to CpG Islands
- Set table to cpgIslandExt
- Set output format to BED
- Set output file to cpg.bed
- Select get output
The Output cpgIslandExt as BED page will open.
- Select get BED to download a compressed folder containing the BED file
- Unzip the file using 7-Zip, WinRAR, or a similar program of your choice to a location you will be able to find
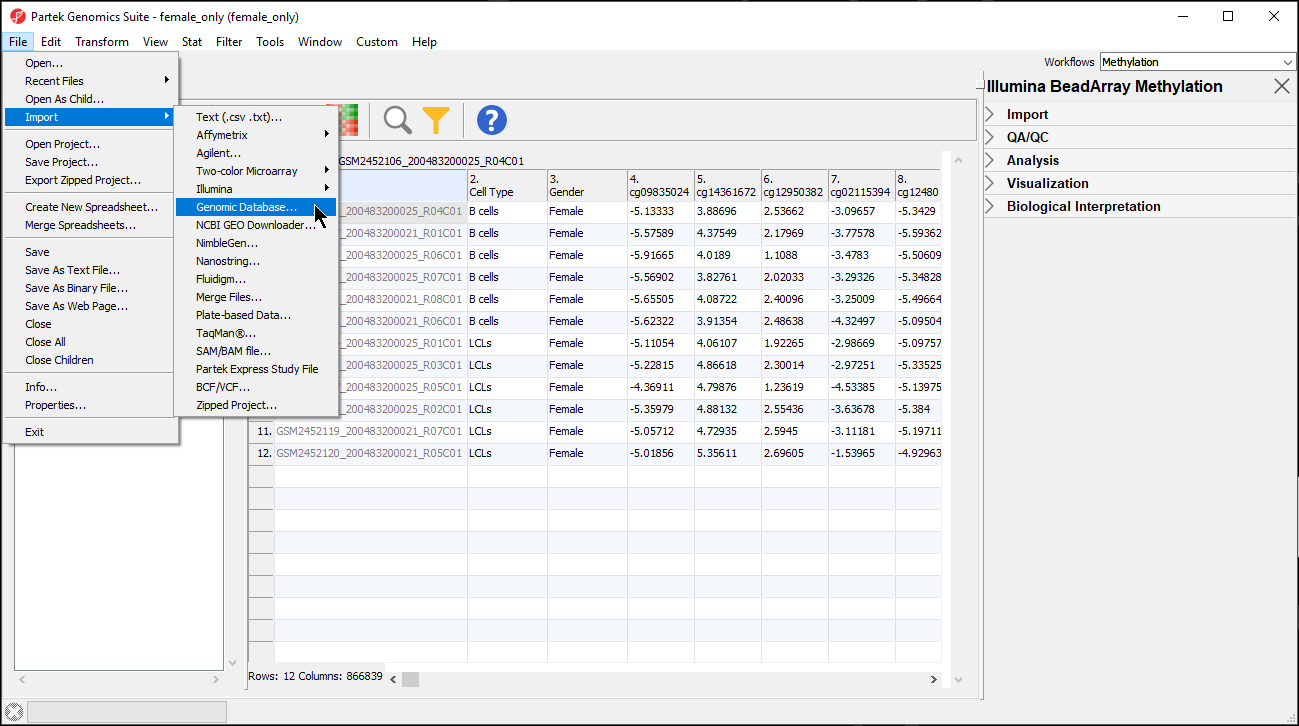
Next, we can import the BED file into Partek Genomics Suite.
- Select Genomic Database... under Import under File in the main toolbar in Partek Genomics Suite (Figure 3)
- Select the file cpg.bed
The BED file will open as a new spreadsheet.
- Change the spreadsheet name to UCSC CpG Island Annotation and save it
For this region list, you can also calculate the average beta values for the probes in each island per sample and detect differential methylated CpG islands regions. Detailed information on how to get average beta value for each CpG can be found in the Determining the average values for a region list section of Starting with a list of genomic regions.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |