Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
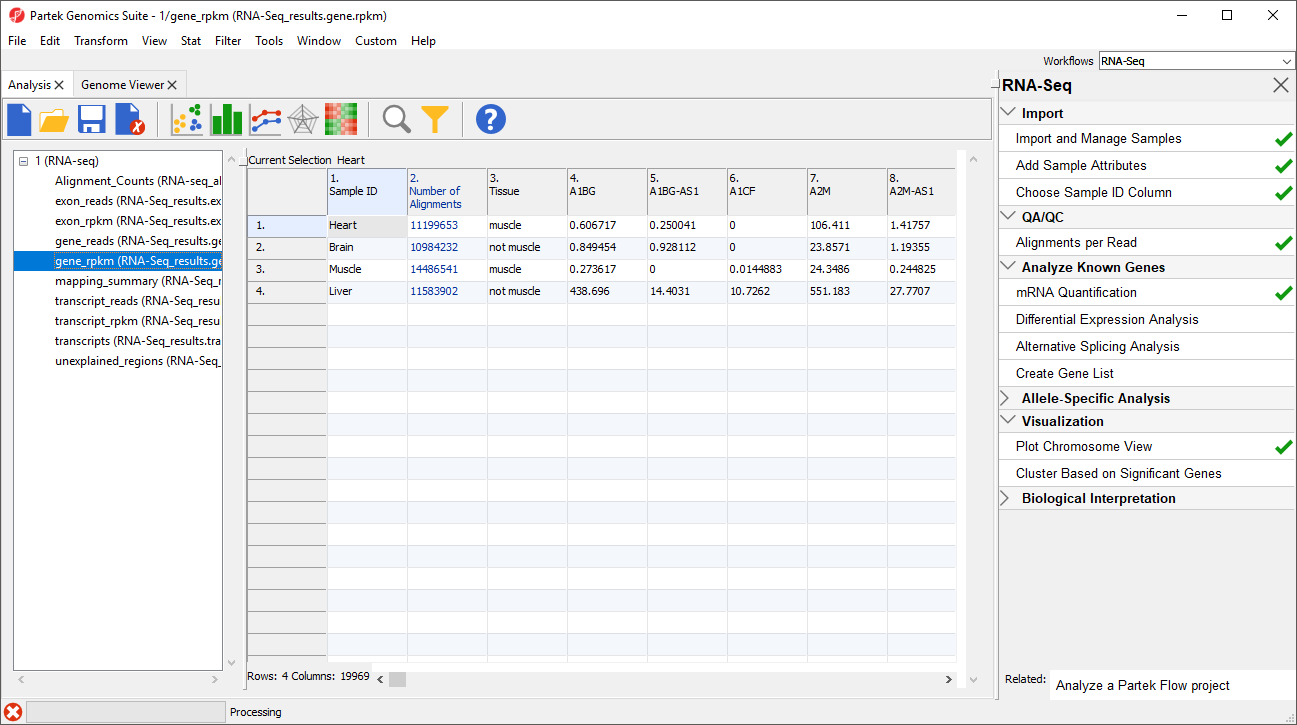
The _reads and _rpkm spreadsheets
Data on features - genes, transcripts, and exons - is presented before and after normalization as _reads and _rpkm spreadsheets. In this tutorial, we have created exon_reads, exon_rpkm, gene_reads, gene_rpkm, transcript_reads, and transcript_rpkm spreadsheets.In these spreadsheets, samples are listed one per row and the normalized counts of the reads mapped to features are in columns (Figure 2).
The _reads and _rpkm spreadsheets can be used for data analysis. Sample grouping can be visualized using PCA. Select View > Scatter Plot from the toolbar or press on the quick action bar to create a PCA plot from the selected spreadsheet. See Exploratory Gene Expression Data Analysis for an example of using PCA plots for data analysis or consult Chapter 7 of the Partek User's Manual for a detailed introduction to PCA. With replicates in a sample group, you would also be able to use the _rpkm perform Differential expression analysis using ANOVA.
The transcripts spreadsheet
The transcripts spreadsheet details the analysis results of RNA-Seq if there are no replicates. Each row lists a separate transcript in each row.
It is possible to derive basic information about from the RNA-Seq_result.transcripts spreadsheet about differential and alternative splicing between your samples even if you don’t have replicates from the RNA-Seq_result.transcripts spreadsheet using a simple chi-squared or log-likelihood tests since because each sample is represented only once and the we can assume a null hypothesis is that the transcripts are evenly distributed across all samples. However, the power of Partek Genomics Suite software resides in the implementation of a mixed-model ANOVA that can handle unbalanced and incomplete datasets, nested designs, numerical and categorical variables, any number of factors, and flexible linear contrasts when you do have biological replicates.
...