...
- Under the Visualization section in the Gene Expression workflow, select Cluster Based on Significant Genes (Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | Select cluster based on significant genes from the Visualization panel of the Gene Expression workflow |
|---|
| AnchorName | Selecting Clustering |
|---|
|
|
...
- The Cluster Significant Genes dialog asks you to specify the type of clustering you want to perform. Select Hierarchical Clustering and select OK
- Choose the Down_Syndrome_vs_Normal (A) spreadsheet under the Spreadsheet with differentially expressed genes
- Choose the Standardize – shift genes to mean of zero and scale to standard deviation of one under the Expression normalization panel. This option will adjust all the gene intensities such that the mean is zero and the standard deviation is 1
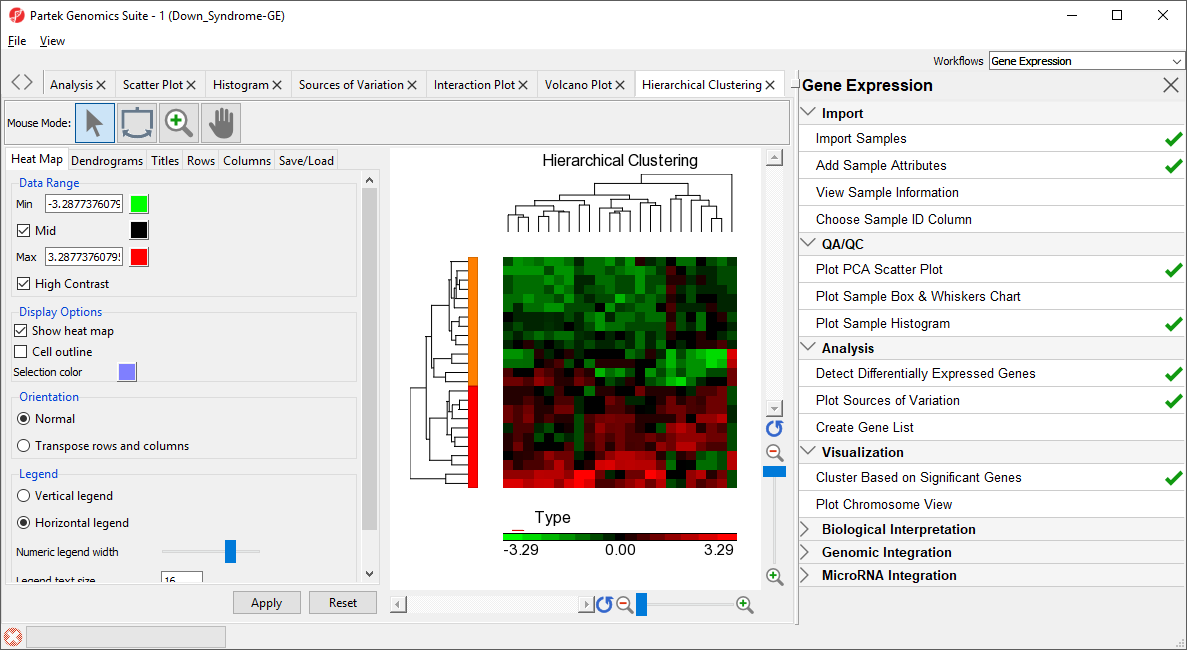
- Select OK to generate a Hierarchical Clustering tab (Figure 222)
| Numbered figure captions |
|---|
| SubtitleText | Hierarchical Clustering of Down_Syndrome_vs_Normal (A) |
|---|
| AnchorName | Hierarchical Clustering |
|---|
|
|
The graph (Figure 222) illustrates the standardized gene expression level of each gene in each sample. Each gene is represented in one column, and each sample is represented in one row. Genes which are unchanged are have a value of zero and are colored black. Genes with increased expression have positive values and are colored red. Genes with reduced expression have negative values and are colored green. Down syndrome samples are colored red and normal samples are colored orange. On the left-hand side of the graph, we can see that the Down syndrome samples cluster together.
...
- In the Down_Syndrome_vs_Normal (A) spreadsheet, right click on the second column header 2. ProbesetID and select Insert Annotation from the pop-up menu (Figure 233)
| Numbered figure captions |
|---|
| SubtitleText | Inserting an annotation |
|---|
| AnchorName | Adding Annotation to Gene List |
|---|
|
|
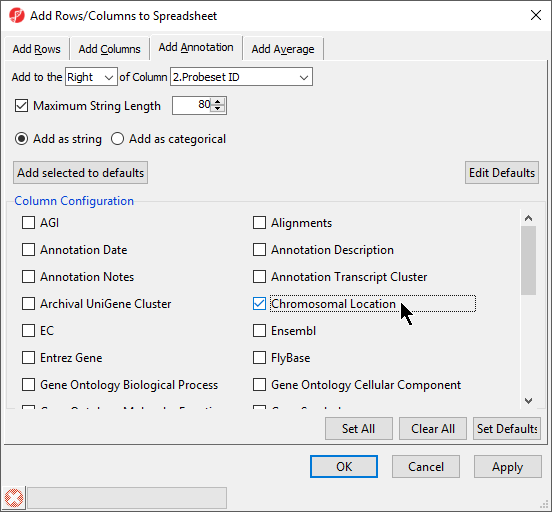
- Select Chromosomal Location under the Column Configuration panel (Figure 4). Leave everything else as default and select OK
| Numbered figure captions |
|---|
| SubtitleText | Adding Chromosomal Location annotation |
|---|
| AnchorName | Add Rows/Columns to Spreadsheet |
|---|
|
|
Interestingly, of the 23 genes of the Down_Syndrome_vs_Normal (A) spreadsheet, 20 genes are located on chromosome 21. To save changes to the spreadsheet, select the Save Active Spreadsheet icon ( Image Added).
Image Added).
- To create a dot plot showing expression levels of a specific gene for each sample, right click on the row header and select Dot Plot (Orig. Data) from the pop-up menu. This generates a Dot Plot tab for the selected gene (Figure 5)
| Numbered figure captions |
|---|
| SubtitleText | Dot Plot showing DSCR3 expression levels for each sample |
|---|
| AnchorName | Dot Plot |
|---|
|
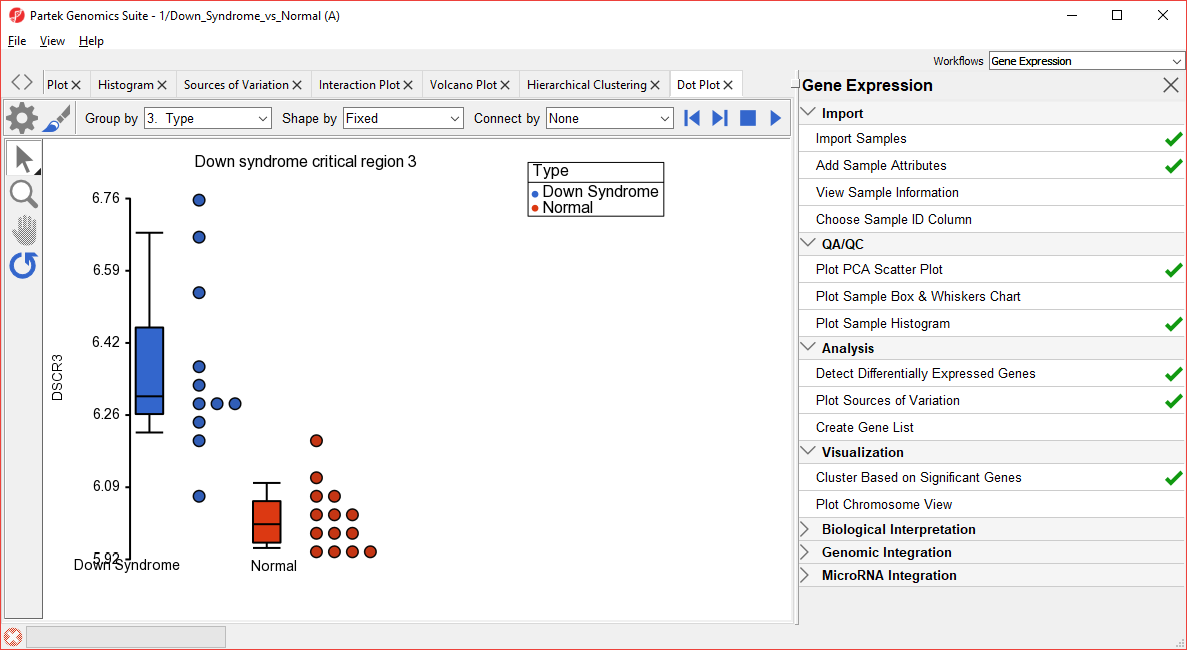 Image Added Image Added
|
In the plot, each dot is a sample of the original data. The Y-axis represents the log2 normalized intensity of the gene and the X-axis represents the different types of samples. The median expression of each group is different from each other in this example. The median of the Down syndrome samples is ~6.3, but the median of the normal samples is ~6.0. The line inside the Box & Whiskers represents the median of the samples in a group. Placing the mouse cursor over a Box & Whiskers plot will show its median and range.




